Abstract
The protein phosphatase 2A complex (PP2A), the major Ser/Thr phosphatase in the brain, is involved in a number of signalling pathways and functions, including the regulation of crucial proteins for neurodegeneration, such as alpha-synuclein, tau and LRRK2. Here, we report the identification of variants in the PTPA/PPP2R4 gene, encoding a major PP2A activator, in two families with early-onset parkinsonism and intellectual disability.
We carried out clinical studies and genetic analyses, including genome-wide linkage analysis, whole-exome sequencing, and Sanger sequencing of candidate variants. We next performed functional studies on the disease-associated variants in cultured cells and knock-down of ptpa in Drosophila melanogaster.
We first identified a homozygous PTPA variant, c.893T>G (p.Met298Arg), in patients from a South African family with early-onset parkinsonism and intellectual disability. Screening of a large series of additional families yielded a second homozygous variant, c.512C>A (p.Ala171Asp), in a Libyan family with a similar phenotype. Both variants co-segregate with disease in the respective families. The affected subjects display juvenile-onset parkinsonism and intellectual disability. The motor symptoms were responsive to treatment with levodopa and deep brain stimulation of the subthalamic nucleus.
In overexpression studies, both the PTPA p.Ala171Asp and p.Met298Arg variants were associated with decreased PTPA RNA stability and decreased PTPA protein levels; the p.Ala171Asp variant additionally displayed decreased PTPA protein stability. Crucially, expression of both variants was associated with decreased PP2A complex levels and impaired PP2A phosphatase activation. PTPA orthologue knock-down in Drosophila neurons induced a significant impairment of locomotion in the climbing test. This defect was age-dependent and fully reversed by L-DOPA treatment.
We conclude that bi-allelic missense PTPA variants associated with impaired activation of the PP2A phosphatase cause autosomal recessive early-onset parkinsonism with intellectual disability. Our findings might also provide new insights for understanding the role of the PP2A complex in the pathogenesis of more common forms of neurodegeneration.
Keywords: PTPA, PPP2R4, parkinsonism, intellectual disability, PP2A
Fevga, Tesson, Carreras Mascaro, Courtin et al. identify homozygous PTPA variants co-segregating with parkinsonism and intellectual disability in two families. Both variants are associated with reduced PTPA levels and impaired activation of PP2A in vitro. PTPA knockdown in Drosophila induces levodopa-sensitive motor impairment.
Introduction
The identification of monogenic forms of Parkinson’s disease (PD) and parkinsonism has shed light on molecular pathways underlying the disease pathogenesis and pointed to new therapeutic targets. Rare heterozygous variants in SNCA,1LRRK2,2,3 and VPS354,5 are established causes of autosomal dominant forms of typical PD, while bi-allelic variants in PRKN,6PINK1,7 and DJ-18 cause autosomal recessive (AR) forms of PD, usually of early-onset. Furthermore, disease-causing variants in several other genes have been shown to cause atypical forms of juvenile parkinsonism, with variable occurrence of additional clinical features. Yet, other causative genes likely remain to be identified, as in many families, particularly with early-onset parkinsonism, variants are not detected in any established/nominated gene.
In this study, we nominate PTPA/PPP2R4 as a novel gene for early-onset parkinsonism and intellectual disability (ID). The encoded PP2A Phosphatase Activator (PTPA) is a major modulator of the protein phosphatase 2A (PP2A) complex function, and it is ubiquitously expressed throughout the human organism and within the brain. PP2A is the major Ser/Thr phosphatase in the brain and targets a myriad of proteins involved in diverse cellular functions such as cellular metabolism,9 cell cycle and proliferation,10 embryonic development,11 cytoskeleton dynamics,12 and apoptosis.13 The PP2A core dimer is composed of a catalytic (PP2A-C) and a scaffolding subunit (PP2A-A). This core dimer associates with one of more than a dozen regulatory subunits (PP2A-B) to form the PP2A heterotrimeric holoenzyme.14 These alternative regulatory subunits confer substrate and (sub)cellular localization specificity to the enzyme.14
The activation of PP2A15,16 is highly dependent on its methylation,17–20 which is negatively regulated by the PP2A-specific methylesterase, PME-1.17,18 PTPA plays a key activating role by binding PME-1 and counteracting its negative influence on PP2A.19,20 This key modulating role of PTPA on PP2A function has also been shown in vivo, where PTPA loss-of-function (LOF) was found to resemble a PP2A-deficient state.16 Furthermore, PTPA deletion has been found to be lethal in fruit fly21 and yeast,16,22,23 further highlighting its essential role in PP2A regulation.
PP2A dysregulation has been implicated in neurodegenerative disorders. The methylated, and therefore active, form of PP2A appears to be decreased in post-mortem brains from subjects with PD, Alzheimer’s disease, dementia with Lewy bodies, and progressive supranuclear palsy, whereas the level of demethylated PP2A is increased.24 Furthermore, PP2A is known to directly target and dephosphorylate alpha-synuclein,25 tyrosine hydroxylase,26 LRRK2,27 and tau.28 PP2A has been shown to dephosphorylate phospho-Ser129 alpha-synuclein, the aggregation-prone form of alpha-synuclein.25 Moreover, enhanced PP2A activity has been demonstrated to reduce phosphorylation and subsequent activation of tyrosine hydroxylase, the rate-limiting enzyme in dopamine synthesis.29,30 Finally, downregulation of PP2A activity might promote tau pathology in Alzheimer’s disease31,32 as well as cognitive and electrophysiological impairments in transgenic mice.32
Interestingly, deregulation of PTPA activity has also been observed in Angelman syndrome, a neurodevelopmental disorder with severe ID due to UBE3A LOF, encoding an E3 ubiquitin ligase that ubiquitinates PTPA.33 To our knowledge, however, variants in PTPA have not previously been described as a cause of monogenic human disease. Here, we report the identification of different bi-allelic variants in PTPA, associated with impaired activation of PP2A, in patients from two families with parkinsonism and ID.
Materials and methods
Patients
The study procedures were approved by the ethical authorities at the participating institutions, and written informed consent was obtained for each participant. Initially, a South African family with two siblings affected by juvenile-onset parkinsonism and ID (Family 1) was clinically characterized and genetically studied. Subsequently, two large independent series of unrelated cases with early-onset (onset <40 years) and/or recessive PD/parkinsonism were studied in order to identify additional PTPA variants. In a first series of 200 patients (Erasmus MC Rotterdam and the International Parkinsonism Genetics Network), the entire coding region and exon–intron boundaries of PTPA (NM_178001) were screened by Sanger sequencing (primers listed in Supplementary Table 1). This series comprised 91 probands with familial PD or parkinsonism, compatible with AR inheritance (12 patients with early-onset), 78 patients with sporadic early-onset PD, and 31 patients with early-onset atypical parkinsonism (four with parkinsonism and ID).
The second series assembled by the French and Mediterranean Parkinson’s Disease Genetics Study group (FMPD cohort) consisted of 589 index PD patients with available whole-exome sequencing (WES) data. Most of these patients present with early-onset PD. In this study group, no variants of interest were identified in known genes causing or related to PD/parkinsonism (Supplementary Table 2).
This cohort included 661 patients with PD, 258 females and 403 males, from 589 families. Four hundred and ninety-two were from Europe, 82 from North Africa, seven from South Africa, and 80 of unknown origin. Among them, 87 families with at least two affected siblings (n = 159 patients) were compatible with AR PD, including 13 families (n = 22 patients) with parental consanguinity. 502 patients were isolated cases, including 62 with consanguinity. This cohort included 41 patients with familial early-onset parkinsonism (30 families) and three patients with early-onset parkinsonism and ID. WES data were available for all index cases, 72 additional affected members and 50 unaffected relatives. The mean age at onset of motor signs was 38.7 years (range: 6–80 years). Seventeen patients presented either MMSE <24, intellectual impairment or cognitive decline. Information on cognitive status was not available for 78 patients.
Genetic studies
Regarding Family 1, genomic DNA was extracted from peripheral blood using standard protocols. High-density genome-wide genotyping in Subjects F1-I-1, F1-I-2, F1-II-1, and F1-II-2 (Fig. 1A) was carried out with the Infinium® CytoSNP-850K v1.1 kit (Illumina), and we performed linkage analysis to generate a list of genomic regions of interest, defined as regions with logarithm of odds (LOD) score >0. Multipoint parametric linkage analysis was performed with MERLIN34 assuming an AR mode of inheritance.
Figure 1.
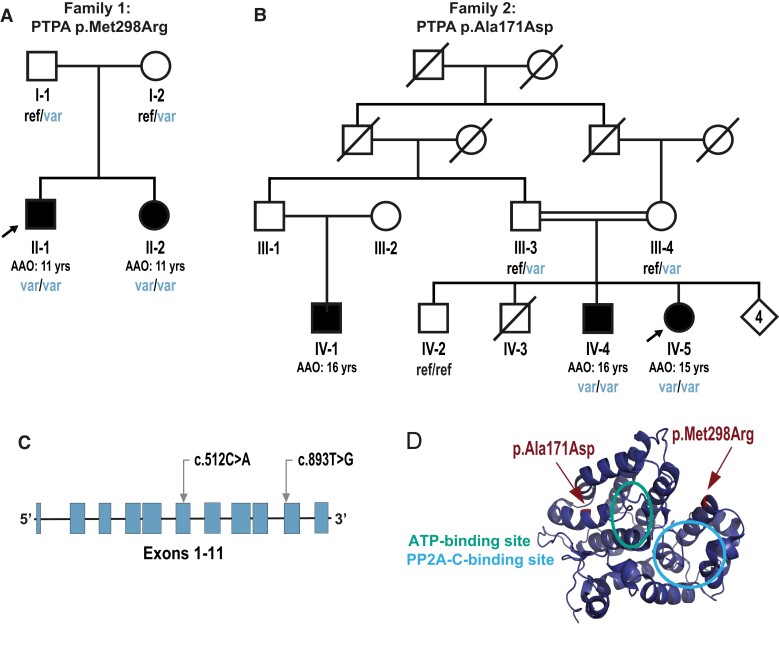
Pedigrees and identified PTPA variants. A and B show the pedigrees of Families 1 and 2, respectively. Filled symbols indicate individuals affected by parkinsonism; unfilled symbols indicate unaffected individuals; arrows denote index cases. AAO = age at onset; ref/var = heterozygous genotype for the corresponding variant; var/var = homozygous variant genotype; ref/ref = wild-type (reference) genotype. (C) PTPA gene schematic representation, including relative positions of the variants identified in this study annotated according to NM_178001. (D) 3D molecular rendering of PTPA, with the relative positions of the variants and the ATP binding and PP2A-C binding sites.
WES on Subject F1-II-2 was performed by using the Nextera Rapid Capture Exome v1.2 kit (Illumina) and HiSeq4000, paired-end 150-bp sequencing (Illumina). The reads were processed according to the GATK v3.7 guidelines,35 and aligned to the human reference genome GRCh37/hg19 with the Burrows-Wheeler Aligner (BWA).36 The variants were called with GATK HaplotypeCaller.35 Finally, WES variants were filtered using Alissa Interpret v5.3 (Agilent Technologies).
In the WES data of Subject F1-II-2, we first searched for variants in genes causing/related to PD/parkinsonism (Supplementary Table 3). We then searched for variants located within the regions of interest with LOD >0 resulting from the AR linkage analysis model. Variants were filtered if they (i) had a minor allele frequency (MAF) <0.1% in public population databases as well as in in-house reference datasets; (ii) were homozygous or double heterozygous; and (iii) had a coding effect or were located within ±5 bp from intron-exon boundaries and predicted to affect splicing by at least one out of four tools (ADA,37 RF,37 SpliceAI,38 and SQUIRLS39). Predictions and scores across in silico algorithms were obtained via the Ensembl Variant Effect Predictor (VEP) v105.40
Furthermore, genome-wide copy number variant analysis was performed with Nexus Copy Number v10 (BioDiscovery), PennCNV,41 and QuantiSNP.42 Copy number variants were considered of interest if they fulfilled the following criteria: called by at least two out of the three above-listed analysis tools and (i) located in genes causing and/or related to PD/parkinsonism (Supplementary Table 3) or (ii) spanning coding genes located in the linkage regions of interest with LOD >0, in bi-allelic state in both affected siblings.
WES in the FMPD cohort was performed at the ICM IGenSeq core facility, Integragen service (Evry, France) or through the NIH facility. Exons were captured using the SureSelect Human All Exon Kits Version 2 Agilent, the Human All Exon V4 + UTRs—70 Mb Agilent, the Human All Exon Agilent, the Roche V.3, Medexome or the Twist Refseq kit, followed by a massively parallel sequencing on the HiSEQ 2000, or the NextSeq500 or NovaSeq system (Illumina). The mean coverage was 104.6× (range 51.1–216.4×) and 25-, 30-, 50-fold mean sequencing depth was achieved across 85.1% (range 79.5–90.4%), 86% (range 58.4–95.5%) and 80.9% (range 57.0–97.3%) of targeted regions, respectively. Read alignment and variant calling were done using an in-house pipeline. Briefly, FastQC was used to check the quality of the reads and low-quality reads were removed using Trimmomatic. Sequencing data were then aligned to the human reference genome hg19 using the bwa suite36 and variant calling was performed using GATK HaplotypeCaller35 or Dragen (Illumina).
Exomes were analysed using VaraFT software.43 Since all known PD genes (Supplementary Table 2) had been previously excluded, we filtered all homozygous or double heterozygous variants in PTPA that possibly affected the cDNA or localized in the splice site region (−8 + 11 bp from exon/intron junction) and with a MAF < 1% in the gnomAD public database.44
To assess whether PTPA variants contribute to the risk of PD, exploratory analyses were also performed in the FMPD cohort and in publicly available PD variant databases (Supplementary material, Appendix 4).
3D structural modelling of PTPA
We generated a schematic representation of the 3D structure of PTPA, including positions of the variants identified in this study as well as positions of ATP binding and PP2A-C binding sites. Atomic coordinates for the PTPA protein were downloaded from Protein Data Bank with accession code 2G62,45 corresponding to the canonical isoform NP_821067 (NM_178000). Functional domains were annotated according to Chao et al.46 Pymol 2.4.1 (Schrödinger LLC) was used to generate the molecular rendering.
Cloning
All DNA constructs used for overexpression experiments were derived from the sequence-verified pCMV3-PTPA cDNA plasmid, which contains the canonical cDNA sequence of PTPA (NM_178000; Sino Biological, HG12287-UT).47 Both variants c.893T>G and c.512C>A were introduced using the QuikChange Lightning site-directed mutagenesis kit (Agilent, 210518), as recommended by the manufacturer. The negative control plasmid was generated by depletion of the PTPA insert via restriction digestion (HindIII and NotI), blunt end generation and T4 ligation. All plasmids were verified by Sanger sequencing.
Cell culture, chemicals and transfection
HEK-293 cell lines were expanded in growth medium (DMEM, ThermoFisher, 10% FCS) at 37°C/5% CO2. Prior to transfection, cells were seeded to a 40–50% confluency in tissue culture plates. Transfections were performed using GeneJuice® reagent (Sigma-Aldrich, 70967) according to manufacturer’s specifications, and the samples were further processed 48 h after transfection. Cycloheximide (40 μg/ml, Merck, C7698-1G), MG-132 (20 µM, Enzo, BML-PI102-0005), Bafilomycin A1 (200 nM, Enzo, BML-CM110-0100) and Torin 1 (200 nM, Invivogen, inh-tor1) were mixed with growth medium and placed on the cells for the indicated time points at 37°C/5% CO2 immediately prior to protein extraction.
Protein extraction
Cultured cells were washed with TBS and lysed with protein lysis buffer (100 mM NaCl, 1.0% IGEPAL® CA-630, 50 mM Tris-Cl, pH 7.4) containing protease inhibitors Complete® (Merck) and Pefabloc® SC (Merck). Lysates were snap frozen, thawed on ice and cleared by centrifugation at 10 000g for 10 min at 4°C.
Western blotting
Protein extracts were mixed with 4× sample buffer (8% SDS, 20% v/v glycerol, 0.002% bromphenolblue, 62.5 mM Tris-Cl, pH 6.8) supplemented with 100 mM dithiothreitol (DTT) and incubated for 10 min at 95°C. Proteins were separated on 4–15% Criterion TGX precast gels (Bio-Rad) and transferred to nitrocellulose membranes using the Trans-Blot® Turbo™ Transfer System (Bio-Rad). Membranes were blocked using 5% non-fat dry milk (Blotto, Santa Cruz Biotechnologies) in TBS, 0.1% v/v Tween® 20 (Merck) for 30 min to 1 h at room temperature. Primary antibody incubations were performed overnight at 4°C in blocking buffer. The following primary antibodies were used: rabbit anti-PTPA (Cell Signaling Technologies, 3330, 1:1000 dilution), mouse anti-PTPA (BioLegend, 824401, 1:1000 dilution), mouse anti-PP2A C (Sigma-Aldrich, 05-421, 1:1000 dilution), rabbit anti-LC3B (Novus, NB100-2220, 1:1000 dilution), rabbit anti-GADD34 (Proteintech, 10449-1-AP, 1:1000 dilution), mouse anti-vinculin (Santa Cruz Biotechnology, V284, 1:2000 dilution). After washing in TBS, 0.1% v/v Tween® 20, blots were incubated for 1 h at room temperature with fluorescently conjugated Alexa Fluor Plus 800 donkey anti-mouse and 700 donkey anti-rabbit (both from ThermoFisher) secondary antibodies. After washing in TBS, 0.1% v/v Tween® 20, blots were imaged and analysed using the Odyssey CLx imaging system (LI-COR Biosciences).
RNA isolation, cDNA generation and RT-qPCR
Total RNA from cultured cells was isolated using the RNeasy Mini Kit (Qiagen) according to manufacturer’s specifications. Integrity and size distribution of total RNA was checked by agarose gel electrophoresis and quantified using a Denovix DS-11_FX spectrophotometer. cDNA synthesis was performed with the RevertAid First Strand Synthesis kit (ThermoFisher, K1622) using 1.5 µg RNA as starting material. RT-qPCR primer sets for targeting PTPA and PP2A-C and reference assays were designed using Primer3 and are listed in Supplementary Table 4.48 Amplifications were carried out in technical triplicates using 0.2 µl of 10× diluted cDNA, 100 nM primers, and 1× TaykonTM No Rox SYBR MasterMix dTTP Blue (Eurogentec, UF-NSMT-B0701) in a final volume of 15 µl. Cycle conditions were 3 min at 95°C followed by 40 cycles of 10 s at 95°C, and 1 min at 60°C in a Bio-Rad CFX96 real-time PCR detection system. Data analysis was performed using Maestro CFX 2.2 software (Bio-Rad). Briefly, the normalized expression of each target gene was calculated using the ΔΔCq method.49 Relative PTPA and PP2A-C mRNA levels were determined after normalization to the geometric mean of the housekeeping genes COPS5 and CLK2.
PP2A phosphatase activity assay
Total protein concentration from transfected HEK-293 cells lysates was determined using the Pierce™ BCA protein assay kit (Thermo Fisher scientific). Protein lysates (100 µg) were mixed with 40 µl of protein A agarose slurry (Santa Cruz Biotechnology, sc-2001) and 2 µl of anti-PP2A C subunit antibody (Sigma-Aldrich, 05-421) in a total of 500 µl with pNPP Ser/Thr assay buffer (Sigma-Aldrich, 20-179), and incubated for 2 h at 4°C with constant rotation. Next, beads were washed three times with TBS and once with pNPP Ser/Thr assay buffer. Subsequently, threonine phosphopeptide (Sigma-Aldrich, 12-219) was added to the washed beads and pNPP Ser/Thr assay buffer to a final concentration of 500 µM as substrate for the enzymatic reaction, and it was incubated for 10 min at 30°C in a shaking incubator. Twenty-five microlitres of the enzymatic reaction were mixed with 100 µl of Malachite Green Phosphate Detection Solution (solution A and additive, Sigma-Aldrich, 20–105 and 20–104, respectively) in a Corning® 96-well half-area microplate (Merck, CLS3695-25EA), and incubated for 10 min at room temperature. Absorbance was measured at a wavelength of 650 nm in a Varioskan microtitre plate reader (ThermoFisher) and compared with the Phosphate standard (Sigma-Aldrich, 20-103).
Drosophila studies
RNA-interference (RNAi) was used for targeted knock-down of gene expression in conjunction with the Gal4/UAS system of cell-type-specific transgene expression. Transgenic RNAi flies were generated based on a previously published protocol.50 Orthologues were identified based on ‘DIOPT ortholog finder’.51 We considered ptpa (FBgn0016698), prkn (FBgn0041100) and nsmase (FBgn0035421) as fly orthologues of PTPA, PRKN and SMPD2, respectively. The transgenic flies were obtained from VDRC Stock Center. The RNAi lines ptpa (VDRC ID: 41912), prkn (VDRC ID: 47636), and nsmase (VDRC ID: 107062) were crossed with a nSyb-Gal4 strain to drive pan-neuronal expression of the RNAi. Flies were raised on a 12:12 light:dark schedule at 25°C and 60% relative humidity. Adult flies of indicated genotypes were tested for behavioural analyses at Days 10, 20, and 35. L-DOPA (D9628, Sigma) was mixed directly into melted fly food at a final concentration of 1 mg/ml. We used prkn as a positive control based on previous studies.52 Based on previous behavioural tests (data not shown), we used nsmase as a negative control. Locomotor activity was evaluated using the climbing test based on negative-geotaxis analysis. The experiment was performed in triplicates, each test including at least 15 flies. The test was performed on all lines five times at each time point after a 1-min rest. Only females were tested. Flies that did not reach the middle of the tube (5 cm) 10 s after stimulation were considered as presenting a locomotor impairment. Fly count was automatically performed by ImageJ.53
Statistical analysis
Statistical analyses were performed using Prism 9 software (GraphPad). For in vitro experiments, one-way ANOVA and Dunnett’s post hoc test were used for experiments in basal conditions and two-way ANOVA followed by Tukey’s post hoc test were used in experiments including drug treatments. Significant P-values of <0.05 or lower were reported. Data are presented as means ± standard deviations (SD) and represent results from three or four biological replicates.
Regarding the Drosophila studies, statistical analyses were performed in R (R 4.1.2, native package). For comparison between two genotypes, the χ2 test was performed.
Data availability
The data that support the findings of this study are available from the corresponding authors, upon reasonable request.
Results
Clinical studies
The clinical features of the patients with PTPA variants are summarized in Table 1, and more detailed case-reports and videotapes of Family 1 patients are available (Supplementary material, Appendix 3, Supplementary Videos 1 and 2). The patients in these two families presented with a very similar phenotype (Table 1 and Supplementary material, Appendix 3). In short, in Family 1 (Fig. 1A) the two affected siblings, born to non-consanguineous parents of South African descent, manifested learning disability and were unable to attend mainstream school in childhood and early adolescence. Early childhood development and motor and communication milestones were delayed in one sibling (Subject F1-II-1) but were normal in the other (Subject F1-II-2). The first signs of parkinsonism manifested at age 11 in both siblings. Brain MRI studies were unremarkable. At the last clinical follow-up (at ages 33 and 24 years, respectively), both patients were still independent in all basic activities of daily life, in keeping with no referred decline in cognitive or language abilities over the years. On the contrary, parkinsonism was clearly progressive in both siblings, with marked positive response to levodopa along with early development of severe, disabling levodopa-induced dyskinesias. Therefore, deep brain stimulation of the subthalamic nucleus (STN-DBS) was carried out, which was effective in both siblings.
Table 1.
Clinical findings in patients with PTPA variants
| Patient/sex | PTPA variant | AAO/AAE | Parkinsonian signs at onset | Other clinical features | Levodopa response | Levodopa-induced dyskinesia | STN-DBS response |
|---|---|---|---|---|---|---|---|
| F1-II-1/male | p.Met298Arg (homozygous) | 11/33 | Rest tremor of the right hand | ID; impulse control disorder while on dopamine agonists; constipation; sialorrhea | + | + | + |
| F1-II-2/female | p.Met298Arg (homozygous) | 11/24 | Gait and upper limbs hypo-/bradykinesia | ID; anxiety; depression; restless legs syndrome | + | + | + |
| F2-IV-5/female | p.Ala171Asp (homozygous) | 15/32 | Bradykinesia; rest tremor of the right hand; rigidity; gait difficulties | ID | + | + | NP |
| F2-IV-4/male | p.Ala171Asp (homozygous) | 16/31 | Bradykinesia | ID | + | - | NP |
- = absent; + = present; AAE = age at last examination; AAO = age at onset of parkinsonian symptomatology; NP = not performed.
The two affected siblings in Family 2 with the PTPA p.Ala171Asp variant were born to healthy first-cousin parents originating from Libya. The index case Subject F2-IV-5 (Fig. 1B), a 32-year-old female, had normal developmental milestones. However, she stopped attending school at age 8 years because of ID. She developed progressive slowness of movements at age 15 years. She noticed insidious appearance of a tremor of the right hand particularly at rest, associated with rigidity of the lower limbs, resulting in gait difficulties. Neurological examination at age 15 years revealed an akinetic-rigid parkinsonian syndrome. Good response to levodopa therapy was obtained initially (500 mg/day) without motor fluctuation. At age 30 years, dyskinesia appeared and persisted despite splitting the daily dose of levodopa into four administrations/day. No sensory, sleep or autonomic dysfunctions, or psychiatric disorders were reported. Neuropsychological evaluation, performed at age 30 years, demonstrated a moderate cognitive impairment [Mini-Mental State Examination (MMSE) = 21]. Brain MRI was normal. Her brother, Subject F2-IV-4 (Fig. 1B), a 31-year-old male, had normal developmental milestones. He stopped attending school at age 7 years due to ID. Since age 16 years, he developed progressive slowness of voluntary movements. There was no evidence of sleep or autonomic dysfunctions or psychiatric disorders. Neurological examination at age 31 years showed an akinetic-rigid parkinsonian syndrome. The administration of levodopa (250 mg/day) induced a complete relief of motor symptoms without dyskinesias. Neuropsychological assessment revealed a moderate cognitive impairment (MMSE = 20). Brain and cervical spinal cord MRI were normal. Their 26-year-old first cousin (Individual F2-IV-1) (Fig. 1B) was followed-up for walking difficulties since age 16 years, displaying progressively worsening symptoms and moderate cognitive impairment. No DNA was available from this patient. Other family members, including both unaffected parents and the 29-year-old unaffected sibling (Individual F2-IV-2), for whom DNA was collected, displayed no signs or symptoms of ID or parkinsonism.
Genetic studies
WES in one sibling from Family 1 (Fig. 1A; Subject F1-II-2) reached an average coverage of 119× of target regions. No variants of interest were identified in known genes causing/related to PD/parkinsonism (Supplementary Table 3). In this family, linkage analysis yielded 41 regions with LOD score >0 (maximum LOD = 0.204) (Supplementary Fig. 1). Filtering these regions for WES variants yielded a unique variant in homozygous state, the PTPA (NM_178001) c.893T>G, p.Met298Arg, and five additional variants in two other genes (Supplementary Table 5). Sanger sequencing confirmed the homozygous PTPA variant in Subject F1-II-2 and co-segregation of this variant with disease in this family (Fig. 1A and Supplementary Fig. 2). Co-segregation analysis excluded all the other variants identified under the AR inheritance model. In addition, we did not find any evidence of copy number variants shared by both affected siblings in this family. The PTPA variant is absent in population databases, predicted deleterious by most in silico algorithms [Combined Annotation Dependent Depletion score (CADD)54 = 32; Table 2 and Supplementary Table 6] and Met298 is highly conserved among orthologues (Supplementary Fig. 3). Taking into account the absence of other disease-co-segregating variants in the genomic regions of interest, the predicted pathogenicity of the PTPA variant, as well as the known functional links between the PP2A complex and neurodevelopmental disorders and neurodegeneration, we considered PTPA as the candidate disease-causing gene in this family.
Table 2.
Homozygous PTPA variants identified in the present study
| Family | Chromosomal positiona | Ref alleleb | Alt allelec | cDNAd | Proteind | gnomAD v2.1.1 | CADDe | GERPe |
|---|---|---|---|---|---|---|---|---|
| Family 1 (South African) |
9:131904725 | T | G | c.893T>G | p.Met298Arg | Absent | 32 | 5.39 |
| Family 2 (Libyan) |
9:131893865 | C | A | c.512C>A | p.Ala171Asp | Absent | 23.6 | 4.94 |
aChromosomal position is given according to GRCh37/hg19.
bRef allele=reference allele.
cAlt allele= alternative allele.
dVariants are annotated based on the PTPA transcript NM_178001.
ePredictions and scores across in silico algorithms were obtained via the Ensembl Variant Effect Predictor (VEP) v10540; CADD_phred_hg1954; GERP= genomic evolutionary rate profiling.55
Screening by means of Sanger sequencing of the first series of 200 unrelated probands with early-onset and/or recessive PD/parkinsonism did not reveal any additional cases with rare homozygous or compound heterozygous variants in PTPA.
To assess the causality of PTPA in the FMPD cohort, we mined WES data from 87 families with AR PD and 502 isolated cases. Variants in PD-associated genes (Supplementary Table 2) were previously excluded. We identified a PTPA homozygous variant in the index case Subject F2-IV-5 (Fig. 1B and Table 2) from a consanguineous Libyan family (Family 2). WES was subsequently performed in the affected brother, both parents and an unaffected brother of the index (Fig. 1B). We searched for homozygous or compound heterozygous variants shared by the two affected siblings, heterozygous in both parents, and either absent or heterozygous in the unaffected sibling. We filtered all variants that possibly affected the cDNA or splice site regions with a MAF < 1% in gnomAD.44 As a result, only the homozygous variant in PTPA, c.512C > A, p.Ala171Asp, remained, which co-segregates with disease in the family, and it was also confirmed by Sanger sequencing (Fig. 1B and Supplementary Fig. 2). This variant, absent in gnomAD,44 is predicted deleterious by many in silico algorithms (CADD54 = 23.6; Table 2 and Supplementary Table 6), and is localized in the 90th percentile constrained coding region.56 Moreover, it affects an amino acid conserved up to zebrafish (Supplementary Fig. 3). Analysis of WES in the remaining 86 families and 502 isolated cases revealed no additional rare homozygous or compound heterozygous variants in PTPA.
Considering both series together, the proportion of unrelated patients that underwent secondary screening and were found to carry bi-allelic PTPA variants was 1/42 (2.38%) for cases with familial early-onset parkinsonism and 1/7 (14.29%) for cases with early-onset parkinsonism and ID.
The PTPA gene is extremely intolerant to LOF variants [pLI = 0.98 for ENST00000337738.1 (NM_178001)],44 suggesting that LOF variants could be pathogenic even in heterozygous state. Therefore, we looked for LOF variants in the FMPD cohort and in the PD variant browser57 (Supplementary material, Appendix 4) but no such variants were found in PD patients therein. Furthermore, we performed burden tests58 using the publicly available data from the PD variant browser57 to explore whether PTPA variants contribute to the risk of developing PD (Supplementary material, Appendix 4). We found no evidence of enrichment of PTPA variants in PD (Supplementary material, Appendix 4).
p.Ala171Asp and p.Met298Arg PTPA variants cause decreased protein and RNA stability
To investigate the effect of the identified disease-associated PTPA variants on PTPA protein and RNA stability, we assessed protein and RNA levels via western blotting and RT-qPCR in an overexpression model.
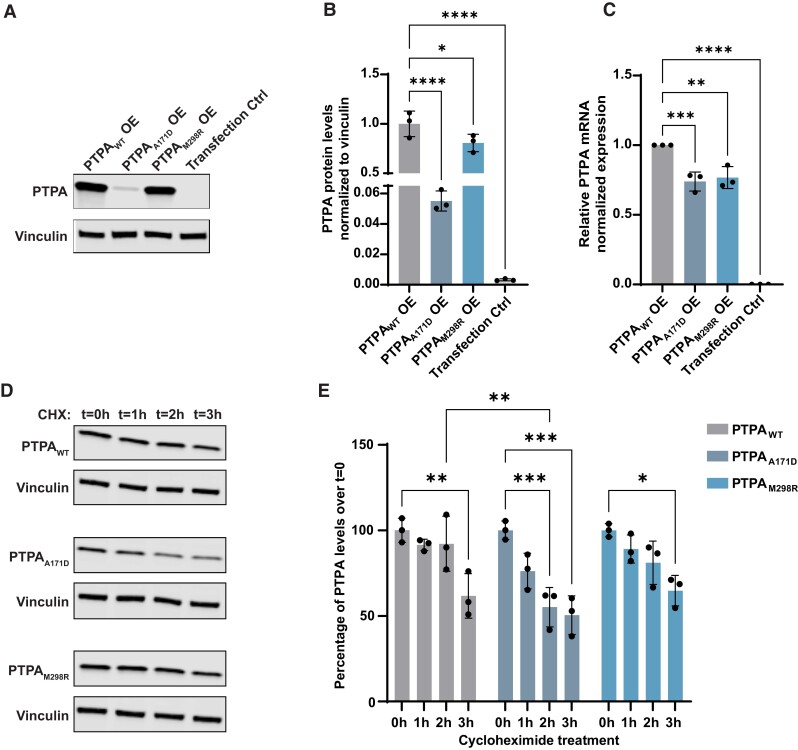
First, we examined PTPA protein levels in cultured cells overexpressing wild-type PTPA (PTPAWT), p.Ala171Asp PTPA (PTPAA171D), p.Met298Arg PTPA (PTPAM298R), and a control plasmid (transfection control). Western blotting (Fig. 2A) revealed a significant decrease in protein levels for both PTPA variants when compared to PTPAWT, with a stronger effect for PTPAA171D (Fig. 2B).
Figure 2.
Comparison of protein and RNA levels of wild-type PTPA and the identified PTPA variants p.Ala171Asp PTPA and p.Met298Arg PTPA in cultured cells. (A) Representative western blot of protein extracts from cultured cells transfected with wild-type PTPA (PTPAWT), p.Ala171Asp PTPA (PTPAA171D), p.Met298Arg PTPA (PTPAM298R) and empty vector (Transfection Ctrl). Blots were probed for expression of PTPA and vinculin. (B) Quantification showing a significant decrease in PTPA levels in p.Ala171Asp and p.Met298Arg PTPA-expressing cells (n = 3). Mean (SD); PTPAWT: 1.00 (0.13); PTPAA171D: 0.06 (0.01); PTPAM298R: 0.81 (0.09); transfection control: 0.00 (0.00). (C) RT-qPCR analysis of PTPA in transfected cells, showing a significant decrease in PTPA expression in PTPAA171D, PTPAM298R and empty vector-expressing cells. Data represent relative normalized expression of PTPA mRNA. CLK2 and COPS5 were used as reference genes (n = 3). Mean (SD); PTPAWT: 1.00 (0.00); PTPAA171D: 0.74 (0.07); PTPAM298R: 0.77 (0.08); transfection control: 0.00 (0.00). (D) Representative western blot of PTPA in transfected cells treated with cycloheximide (CHX) for the indicated times. (E) Quantification of PTPA over cycloheximide treatment, showing a significant decrease in PTPAA171D compared to PTPAWT 2 h after treatment, indicating protein instability. The bars indicate mean PTPA levels per time point as percentage of cells treated at time = 0 h (n = 3). Error bars represent ± SD. Only significant changes *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 are shown.
To assess if RNA stability is involved in the observed reduction of PTPA variant protein levels, we performed RT-qPCR analyses in our overexpression model. We observed reduced relative amounts of PTPA transcript for both variants, indicating RNA instability (Fig. 2C).
Subsequently, to test whether the presence of PTPA variants affected protein stability, we tracked PTPA protein levels over time after blocking protein translation using cycloheximide (Fig. 2C). This experiment revealed that 2 h after treatment, PTPAA171D displayed significantly lower PTPA levels when compared to PTPAWT, while PTPAM298R had a similar turnover rate to PTPAWT, with significantly reduced protein levels 3 h after treatment (Fig. 2D).
Last, we studied PTPA levels upon inhibition of the proteasome and autophagy-lysosome degradation pathways. Treatment for 4 h with MG-132, a potent proteasome inhibitor, led to a significant increase in PTPAA171D levels but had no effect on PTPAWT and PTPAM298R levels (Supplementary Fig. 4A and B). In contrast, PTPAWT and PTPAA171D were sensitive to induction of autophagy via Torin1 combined with lysosomal degradation inhibition via BafA1 for 4 h, whereas PTPAM298R remained unchanged, suggesting an abnormal interaction of this variant with the autophagy-lysosome pathway (Supplementary Fig. 4C and D).
Taken together, these results show evidence for decreased RNA stability for both transcripts encoding PTPAA171D and PTPAM298R variants, and, additionally, decreased PTPA protein stability for PTPAA171D, mainly due to increased proteasomal degradation.
p.Ala171Asp and p.Met298Arg PTPA variants are associated with impaired PP2A complex activation
PP2A complex function is regulated both by post-translational modifications and by altering the levels of its components.59–63 To study the consequences of the expression of the identified PTPA variants on PP2A levels and activity, we studied protein and RNA levels of PP2A-C and quantified the PP2A Ser/Thr phosphatase activity.
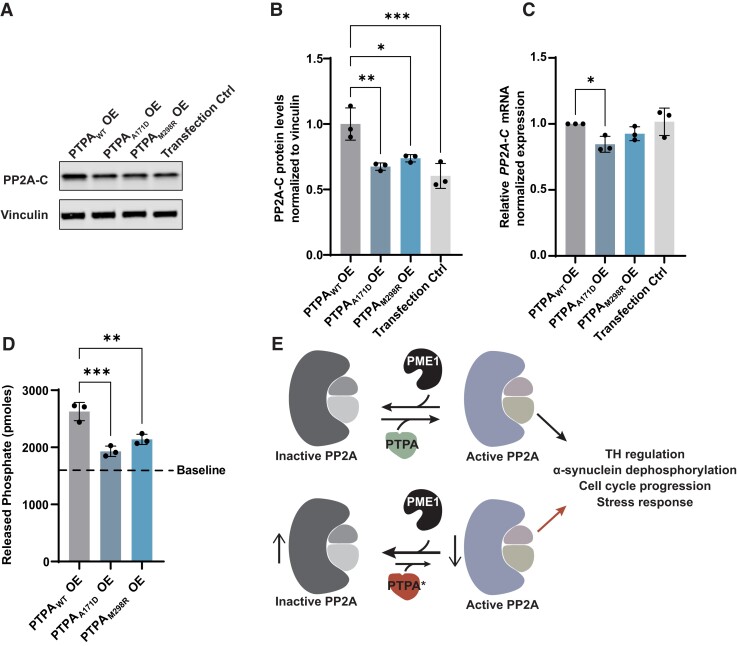
First, comparison of endogenous PP2A-C protein levels by western blotting in cultured cells expressing either PTPAWT or the identified PTPA variants (Fig. 3A) demonstrated that overexpression of PTPAWT leads to a significant increase of endogenous PP2A-C levels, whereas overexpression of each of the PTPA variants does not (Fig. 3B).
Figure 3.
Effect of wild-type PTPA and PTPA variants p.Ala171Asp and p.Met298Arg expression on PP2A complex levels and phosphatase activity. (A) Representative western blot of protein extracts from cultured cells transfected with wild-type PTPA (PTPAWT), p.Ala171Asp PTPA (PTPAA171D), p.Met298Arg PTPA (PTPAM298R) and empty vector (Transfection Ctrl). Blots were probed for expression of PP2A-C and vinculin (B) Quantification showing a significant decrease in PP2A-C levels in PTPAA171D, PTPAM298R and empty vector-expressing cells (n = 3). Mean (SD); PTPAWT: 1.00 (0.12); PTPAA171D: 0.67 (0.03); PTPAM298R: 0.74 (0.03); transfection control: 0.60 (0.09). (C) RT-qPCR analysis of PP2A-C in transfected cells. Quantification shows a mild but significant decrease of PP2A-C levels in PTPAA171D-expressing cells when compared to PTPAWT-expressing cells. Data represent relative normalized expression of PP2A-C mRNA. CLK2 and COPS5 were used as reference genes (n = 3). Mean (SD); PTPAWT: 1.00 (0.00); PTPAA171D: 0.85 (0.06); PTPAM298R: 0.93 (0.05); transfection control: 1.02 (0.10). (D) Quantification of Ser/Thr phosphatase activity assay in transfected cells, showing a significant decrease in activity in PTPAA171D and PTPAM298R-expressing cells (n = 3). Baseline represents pmoles of released phosphate from cells expressing an empty vector. Mean (SD); PTPAWT: 2626 (159.70); PTPAA171D: 1930 (91.24); PTPAM298R: 2139 (90.59); baseline: 1601.67. (E) Scheme representing PTPA-induced activation of PP2A complex phosphatase function by antagonizing the PP2A inhibitor PME-1, and proposed model where the identified PTPA variants p.Ala171Asp and p.Met298Arg are less efficient in inducing PP2A activation. Error bars represent ± SD. Only significant changes *P < 0.05, **P < 0.01 and ***P < 0.001 are shown.
To study the consequence of expression of wild-type or PTPA variants on PP2A-C RNA expression, we performed RT-qPCR in cultured cells. In contrast to what we observed at the protein level, PP2A-C mRNA levels were only mildly affected by PTPA overexpression, with only a significant reduction in cells expressing PTPAA171D (Fig. 3C).
PTPA is a powerful activator of PP2A Ser/Thr phosphatase activity.19,20 We determined the effect of PTPA disease-associated variants on PP2A Ser/Thr phosphatase activity in a cell-free assay by quantifying the amount of released phosphate groups from the substrate by immunoprecipitated PP2A complexes. We observed that expression of the PTPA variants leads to a significant decrease in Ser/Thr phosphatase activity induction when compared to PTPAWT (Fig. 3D). All quantifications were compared to baseline activity in cells expressing endogenous levels of PTPAWT. Taken together, these results indicate that PTPAA171D and PTPAM298R lead to decreased function of PTPA as a PP2A-activator (Fig. 3E).
PTPA loss-of-function induces a locomotor dysfunction in Drosophila that is L-DOPA-reversible
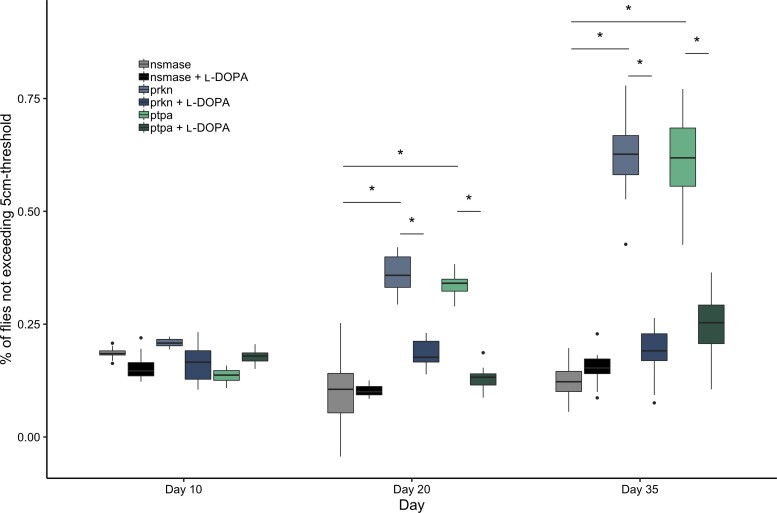
To determine the effect of PTPA LOF in vivo, we turned to Drosophila as a model system. We assayed locomotor activity using the classic negative geotaxis climbing assay and measured performance at different ages upon pan-neuronal RNAi-mediated ptpa knock-down compared to controls; prkn knock-down was used as a positive control and nsmase knock-down as a negative control. No phenotype was observed in 10-day-old flies for any of the lines. At 20 days of age, locomotor dysfunction was observed for the prkn and the ptpa RNAi strains. The proportion of flies that did not reach the 5 cm threshold was significantly higher in both lines compared to nsmase knock-down. Behavioural tests at Day 35 revealed a more severe motor impairment upon knock-down of prkn and ptpa (Fig. 4).
Figure 4.
Effect of ptpa knock-down on D. melanogaster negative geotaxis measured by the climbing test. Comparative climbing test of three different UAS constructs (prkn, ptpa, nsmase) with and without L-DOPA treatment. prkn is used as positive control (significant locomotor impairment). nsmase is used as a negative control (no locomotor impairment). Climbing test performed at Days 10, 20, and 35 represented as proportion of flies that did not reach the 5 cm threshold. No significant difference is observed in locomotor behaviour between ptpa and controls on Day 10. Significant locomotor defect was observed at Days 20 and 35 for the ptpa and prkn lines compared to the negative control nsmase. This defect was significantly corrected with L-DOPA treatment. Significant comparisons are indicated by an asterisk. *P < 0.05. Human and Drosophila orthologues: PRKN (human)/prkn (D. melanogaster); PTPA (human)/ptpa (D. melanogaster); SMPD2 (human)/nsmase (D. melanogaster).
In parallel, we analysed the same lines with food supplemented with L-DOPA. This treatment completely rescued the motor phenotype in the prkn and ptpa lines at Days 20 and 35 (Fig. 4). In contrast, L-DOPA treatment had no effect on locomotor behaviour in the nsmase line.
Taken together, these results suggest that pan-neuronal knock-down of ptpa leads to an age-dependent motor impairment. This defect appears to be dopamine-dependent, as it is reversible with L-DOPA treatment.
Discussion
We present clinical, genetic, and functional studies in two families of South African and Libyan origin with early-onset parkinsonism and ID, associated with variants in the PTPA gene. To our knowledge, this is the first study reporting a monogenic disorder caused by PTPA variants. All four affected homozygotes in this study manifested juvenile-onset parkinsonism and ID. The parkinsonism was progressive, with marked sustained response to levodopa, and in some patients, levodopa-induced dyskinesias and good response to STN-DBS. The intellectual impairment was mild or moderate in these different patients, possibly unmasked with more complex tasks at school, and there was no evidence of marked progression of the cognitive status over time in any of the four patients. Unfortunately, cognitive evaluation using standard rating scales was not available in the siblings in Family 1. However, both siblings had mild ID and they remained independent in basic activities of daily living.
Based on the reported molecular interplay between PP2A and alpha-synuclein, tau, and LRRK2, it is possible that pathological protein aggregation is present in the brains of our patients with PTPA variants. Unfortunately, brain pathology is not available, and whether they have alpha-synuclein, tau and/or other types of protein aggregates remains unknown. Similarly, brain SPECT (DaTSCAN) or PET studies were not available. However, the response to levodopa in both families and STN-DBS in Family 1, as well as the development of levodopa-induced dyskinesias, are in line with the presence of severe presynaptic nigrostriatal dopaminergic deficit.
Further studies in additional patient series and populations will contribute to assess the prevalence of PTPA variants in early-onset PD or parkinsonism with ID, and further characterize the associated phenotypes. Regarding the common forms of late-onset PD, we did not find evidence for enrichment of PTPA variants in PD cases in the PD variant browser,57 and no association of common variants at the PTPA locus with PD is known from the latest meta-analysis of genome wide association studies.64
A variable co-occurrence of parkinsonism and ID has previously been observed in carriers of pathogenic variants in other established or nominated genes, including DNAJC6,65RAB39B,66,67NR4A2/NURR1,68WDR45,69,70 and PTRHD1,71,72 but the neurobiological mechanisms remain incompletely understood. Some of these genes have been implicated in ubiquitous pathways such as vesicular trafficking,67,73–75 autophagy,76–78 and the ubiquitin-proteasome system,79 proposed to be dysfunctional both in parkinsonism/PD and ID.80–85 Future work will determine whether these or other pathways are perturbed in patients with PTPA variants.
Interestingly, rare variants in genes encoding for different subunits/regulators of the PP2A complex have been associated with neurodevelopmental disorders. Patients with PPP2R1A variants present with developmental delay ranging from mild learning difficulties to severe ID,86–88 and other features such as seizures, hypotonia, and hypermobile joints.87 Patients with PPP2CA variants manifest mild to severe ID, developmental delay, behavioural problems, epilepsy, hypotonia, and structural brain abnormalities.89 Variants in other PP2A-related genes, including IGBP1,90,91PPP2R2C,92PPP2R5C,93PPP2R5B,93–95 and PPP2R2B,96 have been described in ID but in some cases the evidence is inconclusive, as they are often found in unique cases or families. More importantly, rare variants in PPP2R5D, previously established in ID and developmental delay with variable occurrence of other clinical features,88,93,97 were recently identified in patients with early-onset levodopa-responsive parkinsonism.93,98–101 Neuropathological examination in a single patient with early-onset parkinsonism, mild ID, developmental delay, and the PPP2R5D p.E200K variant revealed severe neuronal loss and gliosis in the substantia nigra pars compacta with absence of Lewy body pathology.98
Our in vitro studies show several defects in the identified PTPA variants. First, both variants present a reduction in PTPA mRNA levels (Fig. 2C). This reduction cannot be explained by defects during pre-mRNA formation since these experiments were performed using PTPA cDNA lacking the 5′ and 3′ untranslated regions. However, several additional mechanisms have been described where single nucleotide variants lead to decreased RNA stability, and many of these have been associated with diseases, including PD.102 Namely, single nucleotide variants can add or remove binding sites for microRNAs and RNA-binding proteins,103,104 or strongly affect mRNA secondary structures, leading to changes in mRNA levels and functionality.105,106
In addition to decreased PTPA mRNA levels, both variants present lower protein levels (Fig. 2A and B). For the PTPAA171D variant, this effect can be explained by both the lower transcript levels and the decreased protein stability assessed upon cycloheximide treatment (Fig. 2D and E). This increased degradation rate can be potentially explained by more efficient proteasomal degradation (Supplementary Fig. 4A and B), which would be consistent with an abnormal PTPA protein folding due to the presence of p.Ala171Asp. On the other hand, we did not find evidence for protein instability for the PTPAM298R variant, which presented a similar turnover rate to PTPAWT upon cycloheximide treatment (Fig. 2D and E). Therefore, it is possible that the decrease in protein levels for this variant is solely due to lower mRNA levels. Interestingly however, this variant shows a defective interaction with the autophagosomal degradation pathway (Supplementary Fig. 4C and D), suggesting that this variant is being disposed via an alternative proteasome- and lysosome-independent pathway.
Importantly, although the two variants have different effects on PTPA, both lead to a similar outcome, which is the decrease in PP2A phosphatase levels and activity (Fig. 3). While overexpression of PTPAWT leads to increased endogenous PP2A-C protein (but not mRNA) levels, overexpression of PTPAM298R and PTPAA171D fails to do so. PP2A-C levels have been described to be under a complex regulatory control at the translational level, via incompletely understood mechanisms.59–63 However, PP2A-C methylation, which is highly regulated by PTPA, is a well-known dynamic process that determines PP2A-C stability and function.15,16,107 We hypothesize that overexpression of PTPAWT might lead to increased methylation and stability of PP2A-C in PP2A complexes, decreasing its degradation rate; mechanism that is lacking when overexpressing the disease-associated variants, possibly due to decreased binding or activity. Additional work remains ahead to further elucidate the consequences of these variants in the structure of the PP2A complex.
The stimulating role of PTPA on PP2A Ser/Thr phosphatase function is arguably the most relevant function for PTPA.15,16,19 Another function described for PTPA is to stimulate the basal tyrosyl phosphatase activity of PP2A, even though no known endogenous substrates have yet been identified.16,108 It is therefore possible that defects in PTPA impair PP2A function by additional mechanisms. Last, it is unknown whether the identified PTPA variants cause disease by affecting the metabolism/aggregation of alpha-synuclein, tau, tyrosine hydroxylase, or other proteins involved in neurodegeneration, thus further work is warranted. Taken together, our findings from overexpression experiments could indicate that both the identified variants are hypomorphic, and therefore point to PTPA LOF and impaired PP2A stimulation as a common pathogenic mechanism.
Drosophila melanogaster has proven useful for modelling certain aspects of several neurodegenerative disorders, including PD.109–111 Negative geotaxis tested with the climbing assay is a robust behavioural test able to detect subtle changes in flies’ locomotion. Our assays show that neuronal-specific knock-down of the Drosophila PTPA gene orthologue is sufficient to induce a significant locomotor defect in adult mutant flies when compared to the negative control line from Day 20 onwards (Fig. 4). The severity of the phenotype appears to be age-dependent, which might reflect the progressive nature of the underlying neuronal dysfunction or degeneration, similar to the progressive course of parkinsonism in the PTPA variant carriers in this study. This locomotor impairment in ptpa knock-down flies is also dopamine-dependent, as it is fully corrected after L-DOPA treatment, suggesting a possible connection between ptpa expression and the function of dopaminergic neurons. Interestingly, since Drosophila has no homologue of the SNCA gene,109 abnormal phosphorylation levels of other PP2A targets might be responsible for the observed locomotor dysfunction.
To conclude, we report the identification of PTPA as a novel gene associated with juvenile-onset parkinsonism and ID and point to PTPA LOF and impaired activity of the PP2A complex as the most likely pathogenic mechanism for the identified variants. Additional work might provide further insights into the mechanism of the disease caused by PTPA variants, and perhaps to the involvement of PTPA and the PP2A complex in the pathogenesis of more common forms of neurodegeneration.
Supplementary Material
Acknowledgements
We are indebted to the patients participating in this study as well as their families. We thank the DNA and Cell Bank of the ICM Brain Institute for sample preparation, the iGenSeq plateform for exome sequencing and the DAC core facility for bioinformatics analyses. We also thank Pierre Tissier and Aymeric Millecamps for building a device for semi-automated climbing tests. We are grateful to Dr Andrew Singleton for providing some of the WES data through the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services. We acknowledge the support of the DST-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa.
Appendix 1
French and Mediterranean clinicians’ network for Parkinson’s disease genetics (the PDG group) collaborators
Full details are provided in the Supplementary material, Appendix 1.
French PDG collaborators
Yves Agid, Mathieu Anheim, Michel Borg, Alexis Brice, Emmanuel Broussolle, Jean-Christophe Corvol, Philippe Damier, Luc Defebvre, Alexandra Dürr, Franck Durif, Jean Luc Houeto, Paul Krack, Stephan Klebe, Suzanne Lesage, Ebba Lohmann, Maria Martinez, Graziella Mangone, Louise-Laure Mariani, Pierre Pollak, Olivier Rascol, François Tison, Christine Tranchant, Marc Vérin, François Viallet, and Marie Vidailhet.
Collaborators from Mediterranean countries
Ebba Lohmann, Murat Emre, Hasmet Hanagasi, Basar Bilgic, Bedia Marangozoğlu, Mustapha Benmahdjoub, Mohammed Arezki, Sofiane A. Bouchetara, Traki Benhassine, Meriem Tazir, Mouna Ben Djebara, Riadh Gouider, Sawssan Ben Romdhan, Chokri Mhiri, Ahmed Bouhouche.
Appendix 2
Collaborators of the International Parkinsonism Genetics Network
Full details are provided in the Supplementary material, Appendix 2.
Vincenzo Bonifati, Wim Mandemakers, Anneke J. A. Kievit, Agnita J. W. Boon, Joaquim J. Ferreira, Leonor Correia Guedes, Murat Emre, Hasmet A. Hanagasi, Basar Bilgic, Zeynep Tufekcioglu, Bülent Elibol, Okan Doğu, Murat Gultekin, Hsin F. Chien, Egberto Barbosa, Laura Bannach Jardim, Carlos R. M. Rieder, Hsiu-Chen Chang, Chin-Song Lu, Yah-Huei Wu-Chou, Tu-Hsueh Yeh, Leonardo Lopiano, Cristina Tassorelli, Claudio Pacchetti, Cristoforo Comi, Francesco Raudino, Laura Bertolasi, Michele Tinazzi, Alberto Bonizzato, Carlo Ferracci, Roberto Marconi, Marco Guidi, Marco Onofrj, Astrid Thomas, Nicola Vanacore, Giuseppe Meco, Edito Fabrizio, Giovanni Fabbrini, Alfredo Berardelli, Fabrizio Stocchi, Laura Vacca, Paolo Barone, Marina Picillo, Giuseppe De Michele, Chiara Criscuolo, Michele De Mari, Claudia Dell'Aquila, Giovanni Iliceto, Vincenzo Toni, Giorgio Trianni, Valeria Saddi, Gianni Cossu, Maurizio Melis.
Contributor Information
Christina Fevga, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Christelle Tesson, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France.
Ana Carreras Mascaro, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Thomas Courtin, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France; Assistance Publique Hôpitaux de Paris, Hôpital Pitié-Salpêtrière, Département de Génétique, DMU BioGeM, Paris, France.
Riaan van Coller, Department of Neurology, Faculty of Health Sciences, University of Pretoria, Pretoria, South Africa.
Salma Sakka, Research Unit in Neurogenetics, Clinical Investigation Center (CIC) at the CHU Habib Bourguiba, Sfax, Tunisia.
Federico Ferraro, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Nouha Farhat, Research Unit in Neurogenetics, Clinical Investigation Center (CIC) at the CHU Habib Bourguiba, Sfax, Tunisia.
Soraya Bardien, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa; South African Medical Research Council/Stellenbosch University Genomics of Brain Disorders Research Unit, Stellenbosch University, Cape Town, South Africa.
Mariem Damak, Research Unit in Neurogenetics, Clinical Investigation Center (CIC) at the CHU Habib Bourguiba, Sfax, Tunisia.
Jonathan Carr, Division of Neurology, Department of Medicine, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa.
Mélanie Ferrien, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France.
Valerie Boumeester, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Jasmijn Hundscheid, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Nicola Grillenzoni, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France.
Irini A Kessissoglou, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France.
Demy J S Kuipers, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Marialuisa Quadri, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Jean-Christophe Corvol, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France; Assistance Publique Hôpitaux de Paris, Hôpital Pitié-Salpêtrière, Département de Neurologie, Centre d'Investigation Clinique Neurosciences, DMU Neuroscience, Paris, France.
Chokri Mhiri, Research Unit in Neurogenetics, Clinical Investigation Center (CIC) at the CHU Habib Bourguiba, Sfax, Tunisia.
Bassem A Hassan, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France.
Guido J Breedveld, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Suzanne Lesage, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France.
Wim Mandemakers, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
Alexis Brice, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, Sorbonne Université, Paris, France; Assistance Publique Hôpitaux de Paris, Hôpital Pitié-Salpêtrière, Département de Génétique, DMU BioGeM, Paris, France.
Vincenzo Bonifati, Department of Clinical Genetics, Erasmus University Medical Center Rotterdam, Erasmus MC, 3015 GD Rotterdam, The Netherlands.
French and Mediterranean Parkinson disease Genetics Study Group:
Yves Agid, Mathieu Anheim, Michel Borg, Alexis Brice, Emmanuel Broussolle, Jean-Christophe Corvol, Philippe Damier, Luc Defebvre, Alexandra Dürr, Franck Durif, Jean Luc Houeto, Paul Krack, Stephan Klebe, Suzanne Lesage, Ebba Lohmann, Maria Martinez, Graziella Mangone, Louise-Laure Mariani, Pierre Pollak, Olivier Rascol, François Tison, Christine Tranchant, Marc Vérin, François Viallet, Marie Vidailhet, Ebba Lohmann, Murat Emre, Hasmet Hanagasi, Basar Bilgic, Bedia Marangozog lu, Mustapha Benmahdjoub, Mohammed Arezki, Sofiane A Bouchetara, Traki Benhassine, Meriem Tazir, Mouna Ben Djebara, Riadh Gouider, Sawssan Ben Romdhan, Chokri Mhiri, and Ahmed Bouhouche
International Parkinsonism Genetics Network:
Vincenzo Bonifati, Wim Mandemakers, Anneke J A Kievit, Agnita J W Boon, Joaquim J Ferreira, Leonor Correia Guedes, Murat Emre, Hasmet A Hanagasi, Basar Bilgic, Zeynep Tufekcioglu, Bulent Elibol, Okan Dog.u, Murat Gultekin, Hsin F Chien, Egberto Barbosa, Laura Bannach Jardim, Carlos R M Rieder, Hsiu-Chen Chang, Chin-Song Lu, Yah-Huei Wu-Chou, Tu-Hsueh Yeh, Leonardo Lopiano, Cristina Tassorelli, Claudio Pacchetti, Cristoforo Comi, Francesco Raudino, Laura Bertolasi, Michele Tinazzi, Alberto Bonizzato, Carlo Ferracci, Roberto Marconi, Marco Guidi, Marco Onofrj, Astrid Thomas, Nicola Vanacore, Giuseppe Meco, Edito Fabrizio, Giovanni Fabbrini, Alfredo Berardelli, Fabrizio Stocchi, Laura Vacca, Paolo Barone, Marina Picillo, Giuseppe De Michele, Chiara Criscuolo, Michele De Mari, Claudia Dell’Aquila, Giovanni Iliceto, Vincenzo Toni, Giorgio Trianni, Valeria Saddi, Gianni Cossu, and Maurizio Melis
Funding
This work was supported by a grant (SPF-1870) from the Stichting ParkinsonFonds (The Netherlands) to VB; the Fondation pour la Recherche Médicale (FRM, MND202004011718), PTC Therapeutics, the Fondation de France, France-Parkinson Association, la Fédération pour la Recherche sur le Cerveau (FRC) and the French program ‘Investissements d’avenir’ (ANR-10-IAIHU-06) to AB; and grants from the South African Medical Research Council (Self-Initiated Research Grant) and the National Research Foundation of South Africa (Grant Number 129249) to SB.
Competing interests
V.B. receives honoraria from Elsevier Ltd. for serving as co-Editor-in-Chief of Parkinsonism & Related Disorders. He also received speaking honoraria from the International Parkinson and Movement Disorder Society, and honoraria as Chair of the MDS International Congress Program Committee (2019–2021).
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Polymeropoulos MH, Lavedan C, Leroy E, et al. . Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. [DOI] [PubMed] [Google Scholar]
- 2. Paisán-Ruíz C, Jain S, Evans EW, et al. . Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. [DOI] [PubMed] [Google Scholar]
- 3. Zimprich A, Biskup S, Leitner P, et al. . Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. [DOI] [PubMed] [Google Scholar]
- 4. Vilariño-Güell C, Wider C, Ross OA, et al. . VPS35 Mutations in Parkinson disease. Am J Hum Genet. 2011;89:162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zimprich A, Benet-Pagès A, Struhal W, et al. . A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011;89:168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kitada T, Asakawa S, Hattori N, et al. . Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. [DOI] [PubMed] [Google Scholar]
- 7. Valente EM, Abou-Sleiman PM, Caputo V, et al. . Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. [DOI] [PubMed] [Google Scholar]
- 8. Bonifati V, Rizzu P, van Baren MJ, et al. . Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. [DOI] [PubMed] [Google Scholar]
- 9. Tung HY, Alemany S, Cohen P. The protein phosphatases involved in cellular regulation. 2. Purification, subunit structure and properties of protein phosphatases-2A0, 2A1, and 2A2 from rabbit skeletal muscle. Eur J Biochem. 1985;148:253–263. [DOI] [PubMed] [Google Scholar]
- 10. Wlodarchak N, Xing Y. PP2A as a master regulator of the cell cycle. Crit Rev Biochem Mol Biol. 2016;51:162–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee WJ, Kim DU, Lee MY, Choi KY. Identification of proteins interacting with the catalytic subunit of PP2A by proteomics. Proteomics. 2007;7:206–214. [DOI] [PubMed] [Google Scholar]
- 12. Hoffman A, Taleski G, Sontag E. The protein serine/threonine phosphatases PP2A, PP1 and calcineurin: a triple threat in the regulation of the neuronal cytoskeleton. Mol Cell Neurosci. 2017;84:119–131. [DOI] [PubMed] [Google Scholar]
- 13. Van Hoof C, Goris J. Phosphatases in apoptosis: to be or not to be, PP2A is in the heart of the question. Biochim Biophys Acta. 2003;1640(2-3):97–104. [DOI] [PubMed] [Google Scholar]
- 14. Sandal P, Jong CJ, Merrill RA, Song J, Strack S. Protein phosphatase 2A - structure, function and role in neurodevelopmental disorders. J Cell Sci. 2021;134:jcs248187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo F, Stanevich V, Wlodarchak N, et al. . Structural basis of PP2A activation by PTPA, an ATP-dependent activation chaperone. Cell Res. 2014;24:190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fellner T, Lackner DH, Hombauer H, et al. . A novel and essential mechanism determining specificity and activity of protein phosphatase 2A (PP2A) in vivo. Genes Dev. 2003;17:2138–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xing Y, Li Z, Chen Y, et al. . Structural mechanism of demethylation and inactivation of protein phosphatase 2A. Cell. 2008;133:154–163. [DOI] [PubMed] [Google Scholar]
- 18. Kaur A, Westermarck J. Regulation of protein phosphatase 2A (PP2A) tumor suppressor function by PME-1. Biochem Soc Trans. 2016;44:1683–1693. [DOI] [PubMed] [Google Scholar]
- 19. Longin S, Jordens J, Martens E, et al. . An inactive protein phosphatase 2A population is associated with methylesterase and can be re-activated by the phosphotyrosyl phosphatase activator. Biochem J. 2004;380(1):111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stanevich V, Jiang L, Satyshur KA, et al. . The structural basis for tight control of PP2A methylation and function by LCMT-1. Mol Cell. 2011;41:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang F, Huang ZX, Bao H, et al. . Phosphotyrosyl phosphatase activator facilitates localization of Miranda through dephosphorylation in dividing neuroblasts. Development. 2016;143:35–44. [DOI] [PubMed] [Google Scholar]
- 22. Van Hoof C, Janssens V, De Baere I, et al. . The Saccharomyces cerevisiae phosphotyrosyl phosphatase activator proteins are required for a subset of the functions disrupted by protein phosphatase 2A mutations. Exp Cell Res. 2001;264:372–387. [DOI] [PubMed] [Google Scholar]
- 23. Rempola B, Kaniak A, Migdalski A, et al. . Functional analysis of RRD1 (YIL153w) and RRD2 (YPL152w), which encode two putative activators of the phosphotyrosyl phosphatase activity of PP2A in saccharomyces cerevisiae. Mol Gen Genet. 2000;262:1081–1092. [DOI] [PubMed] [Google Scholar]
- 24. Park HJ, Lee KW, Oh S, et al. . Protein phosphatase 2A and its methylation modulating enzymes LCMT-1 and PME-1 are dysregulated in tauopathies of progressive supranuclear palsy and Alzheimer disease. J Neuropathol Exp Neurol. 2018;77:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee KW, Chen W, Junn E, et al. . Enhanced phosphatase activity attenuates α-synucleinopathy in a mouse model. J Neurosci. 2011;31:6963–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saraf A, Oberg EA, Strack S. Molecular determinants for PP2A substrate specificity: charged residues mediate dephosphorylation of tyrosine hydroxylase by the PP2A/B’ regulatory subunit. Biochemistry. 2010;49:986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Drouyer M, Bolliger MF, Lobbestael E, et al. . Protein phosphatase 2A holoenzymes regulate leucine-rich repeat kinase 2 phosphorylation and accumulation. Neurobiol Dis. 2021;157:105426. [DOI] [PubMed] [Google Scholar]
- 28. Sontag E, Nunbhakdi-Craig V, Lee G, et al. . Molecular interactions among protein phosphatase 2A, tau, and microtubules. Implications for the regulation of tau phosphorylation and the development of tauopathies. J Biol Chem. 1999;274:25490–25498. [DOI] [PubMed] [Google Scholar]
- 29. Peng X, Tehranian R, Dietrich P, Stefanis L, Perez RG. Alpha-synuclein activation of protein phosphatase 2A reduces tyrosine hydroxylase phosphorylation in dopaminergic cells. J Cell Sci. 2005;118(15):3523–3530. [DOI] [PubMed] [Google Scholar]
- 30. Lou H, Montoya SE, Alerte TN, et al. . Serine 129 phosphorylation reduces the ability of alpha-synuclein to regulate tyrosine hydroxylase and protein phosphatase 2A in vitro and in vivo. J Biol Chem. 2010;285:17648–17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu R, Zhou XW, Tanila H, et al. . Phosphorylated PP2A (tyrosine 307) is associated with Alzheimer neurofibrillary pathology. J Cell Mol Med. 2008;12:241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gnanaprakash M, Staniszewski A, Zhang H, et al. . Leucine carboxyl methyltransferase 1 overexpression protects against cognitive and electrophysiological impairments in Tg2576 APP transgenic mice. J Alzheimers Dis. 2021;79:1813–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang J, Lou SS, Wang T, et al. . UBE3A-mediated PTPA ubiquitination and degradation regulate PP2A activity and dendritic spine morphology. Proc Natl Acad Sci U S A. 2019;116:12500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. [DOI] [PubMed] [Google Scholar]
- 35. McKenna A, Hanna M, Banks E, et al. . The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. . Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–48.e24. [DOI] [PubMed] [Google Scholar]
- 39. Danis D, Jacobsen JOB, Carmody LC, et al. . Interpretable prioritization of splice variants in diagnostic next-generation sequencing. Am J Hum Genet. 2021;108:2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McLaren W, Gil L, Hunt SE, et al. . The ensembl variant effect predictor. Genome Biol. 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang K, Li M, Hadley D, et al. . PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Colella S, Yau C, Taylor JM, et al. . QuantiSNP: an objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Desvignes JP, Bartoli M, Delague V, et al. . VarAFT: a variant annotation and filtration system for human next generation sequencing data. Nucleic Acids Res. 2018;46(W1):W545–WW53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karczewski KJ, Francioli LC, Tiao G, et al. . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. RCSB PDB - 2G62: Crystal structure of PTPA. Accessed 1 October 2021. https://www.rcsb.org/structure/2g62
- 46. Chao Y, Xing Y, Chen Y, et al. . Structure and mechanism of the phosphotyrosyl phosphatase activator. Mol Cell. 2006;23:535–546. [DOI] [PubMed] [Google Scholar]
- 47. CCDS report for consensus CDS. Report for CCDS6920.1. Accessed 15 May 2021. https://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi?REQUEST=CCDS&DATA=CCDS6920
- 48. Primer 3. Accessed 1 May 2021. https://bioinfo.ut.ee/primer3-0.4.0/
- 49. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 50. Qiao H-H, Wang F, Xu R-G, et al. . An efficient and multiple target transgenic RNAi technique with low toxicity in Drosophila. Nat Commun. 2018;9:4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu Y, Flockhart I, Vinayagam A, et al. . An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sang TK, Chang HY, Lawless GM, et al. . A Drosophila model of mutant human parkin-induced toxicity demonstrates selective loss of dopaminergic neurons and dependence on cellular dopamine. J Neurosci. 2007;27:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Davydov EV, Goode DL, Sirota Met al. . Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010;6(12):e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Havrilla JM, Pedersen BS, Layer RM, Quinlan AR. A map of constrained coding regions in the human genome. Nat Genet. 2019;51:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim JJ, Makarious MB, Bandres-Ciga S, et al. . The Parkinson's disease DNA variant browser. Mov Disord. 2021;36:1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 59. Silverstein AM, Barrow CA, Davis AJ, Mumby MC. Actions of PP2A on the MAP kinase pathway and apoptosis are mediated by distinct regulatory subunits. Proc Natl Acad Sci U S A. 2002;99:4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li X, Scuderi A, Letsou A, Virshup DM. B56-associated protein phosphatase 2A is required for survival and protects from apoptosis in Drosophila melanogaster. Mol Cell Biol. 2002;22:3674–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baharians Z, Schönthal AH. Autoregulation of protein phosphatase type 2A expression. J Biol Chem. 1998;273:19019–19024. [DOI] [PubMed] [Google Scholar]
- 62. Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353(3):417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McCright B, Rivers AM, Audlin S, Virshup DM. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J Biol Chem. 1996;271:22081–9. [DOI] [PubMed] [Google Scholar]
- 64. Nalls MA, Blauwendraat C, Vallerga CL, et al. . Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Köroğlu Ç, Baysal L, Cetinkaya M, Karasoy H, Tolun A. DNAJC6 Is responsible for juvenile parkinsonism with phenotypic variability. Parkinsonism Relat Disord. 2013;19:320–324. [DOI] [PubMed] [Google Scholar]
- 66. Lesage S, Bras J, Cormier-Dequaire F, et al. . Loss-of-function mutations in RAB39B are associated with typical early-onset Parkinson disease. Neurol Genet. 2015;1:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wilson GR, Sim JC, McLean C, et al. . Mutations in RAB39B cause X-linked intellectual disability and early-onset Parkinson disease with α-synuclein pathology. Am J Hum Genet. 2014;95:729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wirth T, Mariani LL, Bergant G, et al. . Loss-of-function mutations in NR4A2 cause dopa-responsive dystonia parkinsonism. Mov Disord. 2020;35:880–885. [DOI] [PubMed] [Google Scholar]
- 69. Haack TB, Hogarth P, Kruer MC, et al. . Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am J Hum Genet. 2012;91:1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Manti F, Panteghini C, Garavaglia B, Leuzzi V. Neurodevelopmental disorder and late-onset degenerative parkinsonism in a patient with a WDR45 defect. Mov Disord Clin Pract. 2022;9:110–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Khodadadi H, Azcona LJ, Aghamollaii V, et al. . PTRHD1 (C2orf79) mutations lead to autosomal-recessive intellectual disability and parkinsonism. Mov Disord. 2017;32:287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kuipers DJS, Carr J, Bardien S, et al. . PTRHD1 Loss-of-function mutation in an African family with juvenile-onset Parkinsonism and intellectual disability. Mov Disord. 2018;33:1814–1819. [DOI] [PubMed] [Google Scholar]
- 73. Mignogna ML, Giannandrea M, Gurgone A, et al. . The intellectual disability protein RAB39B selectively regulates GluA2 trafficking to determine synaptic AMPAR composition. Nat Commun. 2015;6:6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pishvaee B, Costaguta G, Yeung BG, et al. . A yeast DNA J protein required for uncoating of clathrin-coated vesicles in vivo. Nat Cell Biol. 2000;2:958–963. [DOI] [PubMed] [Google Scholar]
- 75. Lee DW, Zhao X, Yim YI, Eisenberg E, Greene LE. Essential role of cyclin-G-associated kinase (Auxilin-2) in developing and mature mice. Mol Biol Cell. 2008;19:2766–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Grimmel M, Backhaus C, Proikas-Cezanne T. WIPI-mediated autophagy and longevity. Cells. 2015;4:202–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hedya SA, Safar MM, Bahgat AK. Hydroxychloroquine antiparkinsonian potential: Nurr1 modulation versus autophagy inhibition. Behav Brain Res. 2019;365:82–88. [DOI] [PubMed] [Google Scholar]
- 78. Niu M, Zheng N, Wang Z, et al. . RAB39B deficiency impairs learning and memory partially through compromising autophagy. Front Cell Dev Biol. 2020;8:598622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ishii T, Funakoshi M, Kobayashi H. Yeast Pth2 is a UBL domain-binding protein that participates in the ubiquitin–proteasome pathway. EMBO J. 2006;25:5492–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lewis Patrick A. Vesicular dysfunction and pathways to neurodegeneration. Essays Biochem. 2021;65:941–948. [DOI] [PubMed] [Google Scholar]
- 81. Merino-Galán L, Jimenez-Urbieta H, Zamarbide M, et al. . Striatal synaptic bioenergetic and autophagic decline in premotor experimental parkinsonism. Brain. 2022;145(6):2092–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zheng Q, Huang T, Zhang L, et al. . Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front Aging Neurosci. 2016;8:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Van Bergen NJ, Guo Y, Al-Deri N, et al. . Deficiencies in vesicular transport mediated by TRAPPC4 are associated with severe syndromic intellectual disability. Brain. 2020;143:112–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ji C, Zhao YG. The BPAN and intellectual disability disease proteins WDR45 and WDR45B modulate autophagosome-lysosome fusion. Autophagy. 2021;17:1783–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cheon S, Dean M, Chahrour M. The ubiquitin proteasome pathway in neuropsychiatric disorders. Neurobiol Learn Mem. 2019;165:106791. [DOI] [PubMed] [Google Scholar]
- 86. Deciphering Developmental Disorders Study . Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lenaerts L, Reynhout S, Verbinnen I, et al. . The broad phenotypic spectrum of PPP2R1A-related neurodevelopmental disorders correlates with the degree of biochemical dysfunction. Genet Med. 2021;23:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Houge G, Haesen D, Vissers LE, et al. . B56δ-related protein phosphatase 2A dysfunction identified in patients with intellectual disability. J Clin Invest. 2015;125:3051–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Reynhout S, Jansen S, Haesen D, et al. . De Novo mutations affecting the catalytic Cα subunit of PP2A, PPP2CA, cause syndromic intellectual disability resembling other PP2A-related neurodevelopmental disorders. Am J Hum Genet. 2019;104:139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Graham JM J, Wheeler P, Tackels-Horne D, et al. . A new X-linked syndrome with agenesis of the corpus callosum, mental retardation, coloboma, micrognathia, and a mutation in the alpha 4 gene at Xq13. Am J Med Genet A. 2003;123A:37–44. [DOI] [PubMed] [Google Scholar]
- 91. Piton A, Redin C, Mandel JL. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet. 2013;93:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Backx L, Vermeesch J, Pijkels E, et al. . PPP2R2C, A gene disrupted in autosomal dominant intellectual disability. Eur J Med Genet. 2010;53:239–243. [DOI] [PubMed] [Google Scholar]
- 93. Loveday C, Tatton-Brown K, Clarke M, et al. . Mutations in the PP2A regulatory subunit B family genes PPP2R5B, PPP2R5C and PPP2R5D cause human overgrowth. Hum Mol Genet. 2015;24:4775–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mohrmann I, Gillessen-Kaesbach G, Siebert R, Caliebe A, Hellenbroich Y. A de novo 0.57 Mb microdeletion in chromosome 11q13.1 in a patient with speech problems, autistic traits, dysmorphic features and multiple endocrine neoplasia type 1. Eur J Med Genet. 2011;54:e461–e464. [DOI] [PubMed] [Google Scholar]
- 95. Boyle MI, Jespersgaard C, Nazaryan L, et al. . Deletion of 11q12.3-11q13.1 in a patient with intellectual disability and childhood facial features resembling Cornelia de Lange syndrome. Gene. 2015;572:130–134. [DOI] [PubMed] [Google Scholar]
- 96. Hamdan FF, Srour M, Capo-Chichi JM, et al. . De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shang L, Henderson LB, Cho MT, et al. . De novo missense variants in PPP2R5D are associated with intellectual disability, macrocephaly, hypotonia, and autism. Neurogenetics. 2016;17:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kim CY, Wirth T, Hubsch C, et al. . Early-onset parkinsonism is a manifestation of the PPP2R5D p.E200K mutation. Ann Neurol. 2020;88:1028–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Walker IM, Riboldi GM, Drummond P, et al. . PPP2R5D genetic mutations and early-onset parkinsonism. Ann Neurol. 2021;89:194–195. [DOI] [PubMed] [Google Scholar]
- 100. Hetzelt K, Kerling F, Kraus C, et al. . Early-onset parkinsonism in PPP2R5D-related neurodevelopmental disorder. Eur J Med Genet. 2021;64:104123. [DOI] [PubMed] [Google Scholar]
- 101. Ning P, Li K, Ren H, et al. . Rare missense variants in the PPP2R5D gene associated with Parkinson's disease in the Han Chinese population. Neurosci Lett. 2022;776:136564. [DOI] [PubMed] [Google Scholar]
- 102. Manning KS, Cooper TA. The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol. 2017;18:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ghanbari M, Franco OH, de Looper HW, et al. . Genetic variations in MicroRNA-binding sites affect MicroRNA-mediated regulation of several genes associated with cardio-metabolic phenotypes. Circ Cardiovasc Genet. 2015;8:473–486. [DOI] [PubMed] [Google Scholar]
- 104. Wang G, van der Walt JM, Mayhew G, et al. . Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wan Y, Qu K, Zhang QC, et al. . Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Duan J, Wainwright MS, Comeron JM, et al. . Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–216. [DOI] [PubMed] [Google Scholar]
- 107. Sents W, Meeusen B, Kalev P, et al. . PP2A inactivation mediated by PPP2R4 haploinsufficiency promotes cancer development. Cancer Res. 2017;77:6825–6837. [DOI] [PubMed] [Google Scholar]
- 108. Cayla X, Goris J, Hermann J, et al. . Isolation and characterization of a tyrosyl phosphatase activator from rabbit skeletal muscle and Xenopus laevis oocytes. Biochemistry. 1990;29:658–667. [DOI] [PubMed] [Google Scholar]
- 109. Xiong Y, Yu J. Modeling Parkinson's disease in Drosophila: what have we learned for dominant traits? Front Neurol. 2018;9:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Vanhauwaert R, Verstreken P. Flies with Parkinson's disease. Exp Neurol. 2015;274(Pt A):42–51. [DOI] [PubMed] [Google Scholar]
- 111. Feany MB, Bender WW. A drosophila model of Parkinson's disease. Nature. 2000;404:394–398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors, upon reasonable request.