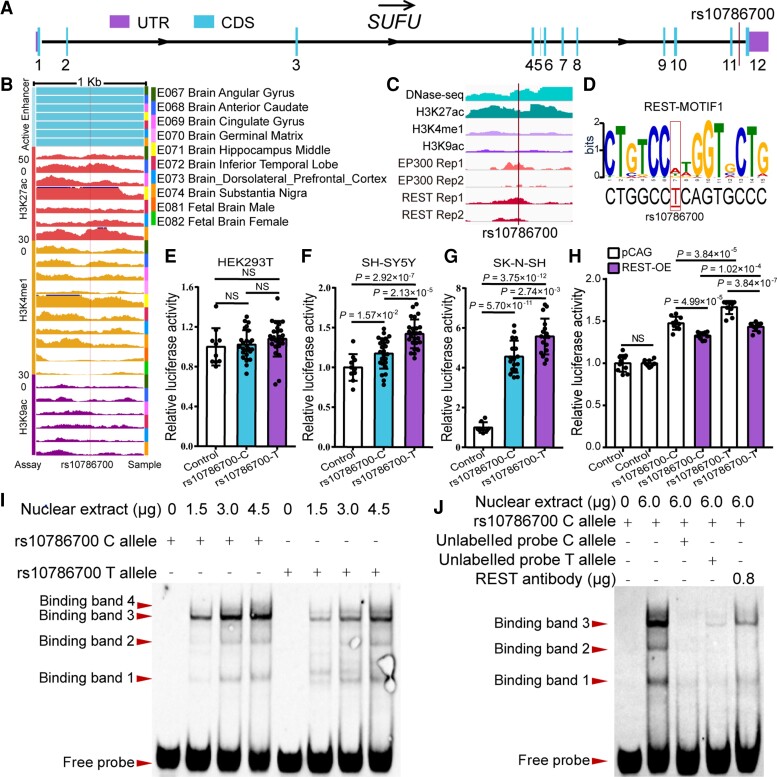
Figure 2.
Validation of the enhancer regulatory effect of rs10786700. (A) rs10786700 is located in the last intron of the Sufu gene. (B) rs10786700 is located in an enhancer element (marked with H3K27ac, H3K4me1 and H3K9ac signals) in human foetal brain. Data were from Roadmap (http://www.roadmapepigenomics.org/). (C) rs10786700 is located in an open chromatin region (i.e. actively transcribed) marked with strong DNase-seq and histone H3K27ac signals in human neuroblastoma cells (SK-N-SH). (D) Position weight matrix (PWM) and FIMO analyses showed that rs10786700 affects REST binding in SK-N-SH cells. (E–G) Dual-luciferase reporter gene assays validated the enhancer activity of rs10786700. The genomic sequence containing rs10786700 exhibited enhancer activity compared with control vector in SH-SY5Y and SK-N-SH cells (but not HEK293T). Of note, the risk allele (C allele) of rs10786700 conferred significant lower luciferase activity compared with the T allele in SH-SY5Y and SK-N-SH cells. Values on the y-axis represent relative reporter gene activity (i.e. the luciferase activity of pGL3 promoter vector containing the cloned sequence relative to the pRL-TK (internal control) vector). n = 8 for control group, n = 16 for experimental groups. (H) REST overexpression significantly repressed the luciferase activity of the genomic fragment containing rs10786700. n = 8. (I) EMSA showed the preferential binding of nuclear extracts to C allele of rs10786700. The loaded amounts of nuclear extracts were 0, 1.5, 3.0 and 4.5 μg, respectively (from left to right). Four binding bands were detected, and the risk allele (C allele) of rs10786700 shows stronger binding affinity to nuclear extracts than the T allele for binding band 3. (J) Super-shift and competitive experiments for EMSA. The binding specificity of rs10786700 to transcription factors was verified by competitive experiments (using unlabelled probes). Super shift experiments were performed to detect the binding of rs10786700 to REST by adding 0.8 μg REST antibody; 6.0 μg total nuclear extracts were added to each lane. The amount of labelled probe was 50 fmol per lane. (I and J) Unpaired two-tailed Student’s t-test; data are presented as mean ± SD; NS, not significant.