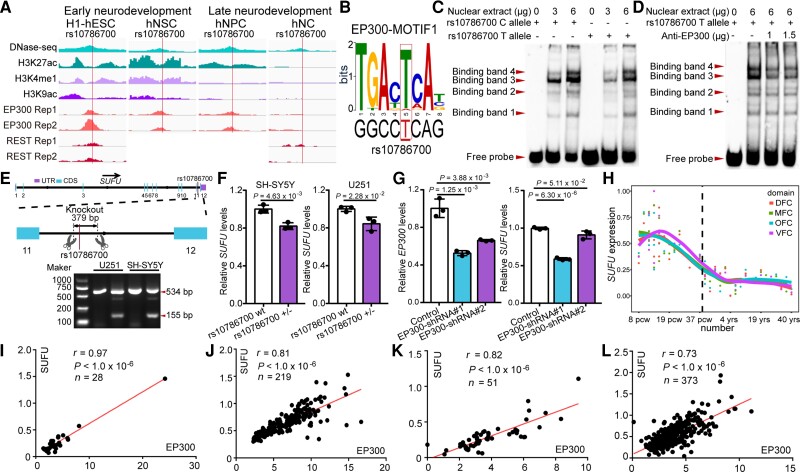
Figure 3.
rs10786700 regulates SUFU expression by dynamically modulating the activity of the SE in neurodevelopment. (A) DNase-seq and histone modifications for genomic region surrounding rs10786700 (1 Mb centred on rs10786700) showed that rs10786700 is located in an actively transcribed genomic region. Of note, ChIP-Seq data showed that EP300 and REST bind to the genomic sequence containing rs10786700 in a development-dependent manner. In early neurodevelopment stage, the ChIP-seq signals of EP300 and REST were high. As development progresses, the ChIP-seq signals decrease gradually. (B) Position weight matrix (PWM) and FIMO analyses showed that rs10786700 disrupts binding of EP300 in SK-N-SH cells. (C) The amounts of nuclear extracts were 0, 3 and 6 μg, respectively (from left to right). Four binding bands were observed. The risk allele (C allele) of rs10786700 showed stronger binding affinity to nuclear extracts than the T allele for binding band 3. (D) Super-shift experiments validated the binding of rs10786700 to EP300. (E) A 379 bp fragment containing rs10786700 (located in the 11th intron of SUFU gene) was knocked out by CRISPR/Cas9-mediated editing in SH-SY5Y and U251 cells. (F) qPCR showed that rs10786700 knock-out resulted in significant downregulation of SUFU expression in both SH-SY5Y and U251 cells. (G) qPCR validation of SUFU expression in EP300 knocked-down SH-SY5Y cells. n = 3 for F and G. Unpaired two-tailed Student’s t-test; data are presented as mean ± SD. (H) Expression patterns of SUFU in the developing and adult human frontal cortex. Expression level of SUFU across the entire developing stages (from 8 pcw to 40 years) were depicted in different areas of the frontal cortex. The expression data (42 human subjects) were from BrainSpan (http://www.brainspan.org/).50 Pcw, post-conception weeks; yrs, years; DFC, dorsolateral prefrontal cortex; MFC, medial prefrontal cortex; OFC, orbital prefrontal cortex; VFC, ventrolateral prefrontal cortex. (I–L) The Pearson expression correlations between SUFU and EP300 in the human brain. (I) Correlations in prenatal human brain (LIBD dataset). (J) Correlations in prenatal human brain (expression data from Walker et al.27). (K) Correlations in childhood human brain. (L) Correlations in adulthood human brain. Sample group: prenatal (age < 0), child (0 < age < 18), adult (age > 18), non-prenatal (age > 0).