Abstract
Glioblastoma is characterized by diffuse infiltration into the surrounding tissue along white matter tracts. Identifying the invisible tumour invasion beyond focal lesion promises more effective treatment, which remains a significant challenge. It is increasingly accepted that glioblastoma could widely affect brain structure and function, and further lead to reorganization of neural connectivity. Quantifying neural connectivity in glioblastoma may provide a valuable tool for identifying tumour invasion.
Here we propose an approach to systematically identify tumour invasion by quantifying the structural connectome in glioblastoma patients. We first recruit two independent prospective glioblastoma cohorts: the discovery cohort with 117 patients and validation cohort with 42 patients. Next, we use diffusion MRI of healthy subjects to construct tractography templates indicating white matter connection pathways between brain regions. Next, we construct fractional anisotropy skeletons from diffusion MRI using an improved voxel projection approach based on the tract-based spatial statistics, where the strengths of white matter connection and brain regions are estimated. To quantify the disrupted connectome, we calculate the deviation of the connectome strengths of patients from that of the age-matched healthy controls. We then categorize the disruption into regional disruptions on the basis of the relative location of connectome to focal lesions. We also characterize the topological properties of the patient connectome based on the graph theory. Finally, we investigate the clinical, cognitive and prognostic significance of connectome metrics using Pearson correlation test, mediation test and survival models.
Our results show that the connectome disruptions in glioblastoma patients are widespread in the normal-appearing brain beyond focal lesions, associated with lower preoperative performance (P < 0.001), impaired cognitive function (P < 0.001) and worse survival (overall survival: hazard ratio = 1.46, P = 0.049; progression-free survival: hazard ratio = 1.49, P = 0.019). Additionally, these distant disruptions mediate the effect on topological alterations of the connectome (mediation effect: clustering coefficient −0.017, P < 0.001, characteristic path length 0.17, P = 0.008). Further, the preserved connectome in the normal-appearing brain demonstrates evidence of connectivity reorganization, where the increased neural connectivity is associated with better overall survival (log-rank P = 0.005).
In conclusion, our connectome approach could reveal and quantify the glioblastoma invasion distant from the focal lesion and invisible on the conventional MRI. The structural disruptions in the normal-appearing brain were associated with the topological alteration of the brain and could indicate treatment target. Our approach promises to aid more accurate patient stratification and more precise treatment planning.
Keywords: brain connectome, glioblastoma, tumour invasion, brain reorganization, survival analysis
Wei et al. characterize topological changes in the connectome in patients with glioblastoma. Distant connectome disruptions—invisible on conventional MRI—are widespread beyond the focal lesions and are associated with impaired cognition and survival. Such changes could have prognostic value in patients with glioblastoma.
Introduction
Glioblastoma is the most common primary malignant brain tumour in adults, characterized by diffuse infiltration into the surrounding tissue.1 It is increasingly accepted that glioblastoma widely influences the brain structure and function beyond the focal lesion.2,3,4,5 Evidence shows that glioblastoma can induce profound reorganization of neural connectivity,5,6 while neuronal activity can promote tumour progression.7 This bidirectional interaction underscores the promise of characterizing neural connectivity for better understanding glioblastoma invasion, which may facilitate more accurate patient stratification for personalized management.
Diffusion-weighted MRI (dMRI) is a method to estimate the structural connectivity of the brain. It is more sensitive to detecting occult tumour invasion than conventional T1-weighted and fluid-attenuated inversion recovery (FLAIR) images.8 Evidence shows that dMRI can indicate the tissue signature of glioma,9 offering value to evaluate invasiveness,10,11 detect peritumoral invasion,12,13 indicate subventricular zone involvement14 and predict molecular phenotypes.15 These studies, however, have focused on the focal tumour, instead of the systematic disturbance of the brain.
The advance in neuroimaging has represented structural connectivity as a complex network, namely structural connectome.16,17 This approach models brain regions as nodes while the white matter (WM) connections among brain regions as edges. Graph theoretical analysis of the derived structural networks shows to characterize various neurological and psychiatric disorders.18,19,20 Moreover, studies suggest that brain tumours may alter the connectome topology while brain networks correspondingly demonstrate robustness and reorganization.5–21,22,23 Recent evidence shows that the topological features derived from the connectome may predict tumour location frequency in glioma patients24 and appear to predict patient survival.25 However, it remains largely unknown whether the connectome disruption could be quantified for patient stratification. Of particular significance is whether the robustness of the connectome could affect patient outcome.
The purpose of this study was to characterize the disruption of the structural connectome in glioblastoma patients and investigate the clinical significance. We hypothesized that glioblastoma could induce both focal and global disturbance to the structural connectome, leading to topological alteration of the brain and impacting patient outcomes. We tested this hypothesis in two prospective glioblastoma cohorts. First, we constructed the structural networks using the dMRI from glioblastoma patients and healthy controls. Second, we quantified the focal and distant disrupted connectome separately from each patient. Third, we calculated the disruption indices and topological features and examined their significance in survival models. Last, we modelled the alteration of the preserved connectivity after removing the disrupted connectome and investigated its significance on patient survival. The results revealed widespread disruptions of the structural connectome, which could lead to topological alterations and show prognostic value in glioblastoma patients.
Materials and methods
Subjects
Patients with a radiological diagnosis of de novo supratentorial glioblastoma were prospectively recruited for resection (discovery: July 2010–August 2015; validation: July 2017–October 2019) by the multidisciplinary team central review. For both cohorts, patients were consecutively recruited following identical inclusion and exclusion criteria, with data prospectively collected (see Supplementary methods). All patients underwent preoperative 3D magnetization-prepared rapid acquisition gradient echo (MPRAGE) [pre-contrast (T1) and post-contrast (T1C)], T2-weighted FLAIR and dMRI sequences. Patient preoperative cognitive performance was tested using the Mini-Mental State Examination (MMSE), dichotomized as <27 or ≥27 as reported.26
We included a control cohort with dMRI and T1 sequences available from https://brain-development.org/ixi-dataset/. We also included the high angular resolution dMRI from the Alzheimer’s Disease Neuroimaging Initiative (ADNI, http://adni.loni.usc.edu/) for constructing an unbiased template of WM connection with high spatial resolution.
Treatment
All patients underwent maximal safe surgery using 5-aminolevulinic acid fluorescence (5-ALA, Medac) and neuro-navigation (StealthStation, Medtronic). For maximal safe resection in both study cohorts, where appropriate, other adjuvants, including awake surgery, cortical and subcortical mapping and intraoperative electrophysiology were also applied. According to the postoperative MRI within 72 h, the extent of resection was assessed as complete or partial resection of enhancing tumour or biopsy. Adjuvant therapy was determined by the multidisciplinary team as standard based on patient postoperative status. All patients were followed up according to the response assessment in neuro-oncology criteria. Overall survival (OS) and progression-free survival (PFS) were used as endpoints.
Tumour segmentation
All anatomical MRI (T1, T2 and FLAIR) were co-registered to T1C images with an affine transformation, using the linear image registration tool (FLIRT) function in the FMRIB Software Library (FSL).27 To segment the tumour, we applied a multi-scale 3D Deep Convolutional Neural Network,28 implemented in the Cancer Imaging Phenomics Toolkit (CaPTk, https://cbica.github.io/CaPTk/index.html). The manual correction was performed using 3D slicer v.4.6.2 (https://www.slicer.org/) by a neurosurgeon (C.L.) and a researcher (Y.W.) after an initial training period and reviewed by an experienced neuroradiologist (T.M.). The final consensus was achieved to ensure inter-rater reliability.
Quantifying the brain connectome
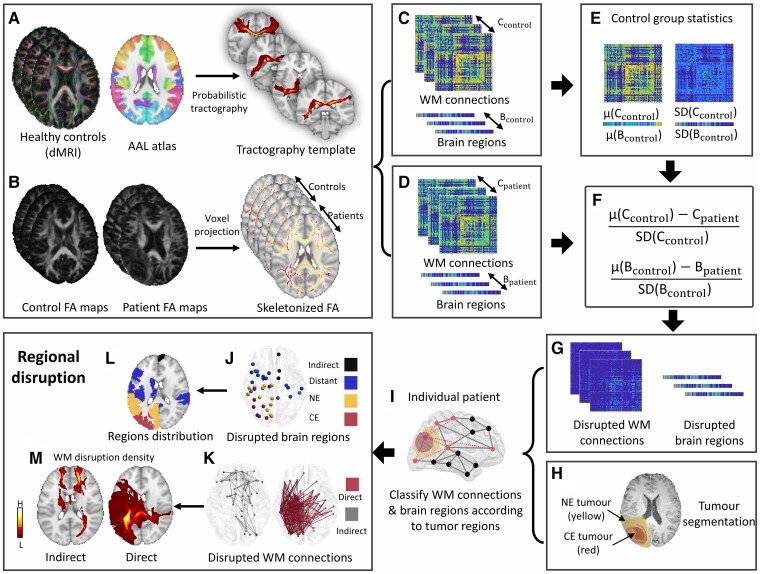
Tractography is a technique to measure the strength of WM connection by tracking the fibre pathway connecting brain regions. However, directly performing tractography on the brain with tumours may cause tracking failure or artefacts, e.g. an unrealistic fibre belt surrounding the tumour.29 To bypass the bias of performing tractography on the lesioned brain, we used an approach to generate a template from healthy controls for tract localization and leveraged the voxel project step of tract-based spatial statistics (TBSS)30 to robustly estimate the strength of WM connection in patients by comparing patients to healthy controls.31,32 The complete pipeline includes the following steps: (i) constructing a group-based tract template (Fig. 1A); (ii) producing individualized skeletonized fractional anisotropy (FA) maps (Fig. 1B); (iii) combining the template with the FA skeleton maps to estimate the connectome strength in both patients and controls (Fig. 1C and D); and (iv) identifying the significantly disrupted connectome in patients (Fig. 1E–G).
Figure 1.
Study design. (A) Probabilistic tractography is performed on the high-resolution dMRI of 10 healthy controls to generate a template for the WM connections among the 90 regions on the Automatic Anatomical Labelling atlas. (B) Skeletonized fractional anisotropy (FA) maps are generated from both patients and age-matched healthy controls to estimate the WM connection strength, using an improved voxel projection procedure based on TBSS. (C and D) The strengths of WM connections are derived in healthy controls and patients by combining WM connection template and skeletonized FA. The strengths of brain regions are calculated by aggregating the strengths of linking WM connections. (E–G) By comparing patients to controls, the disrupted connectome in patients is identified as the WM connection (Cpatient) or brain region (Bpatient) with a strength of over 2SD (95% confidence interval) lower than the mean strength of the control group [μ(Ccontrol) or μ(Bcontrol)]. The disruption indices of WM connections and brain regions are calculated by averaging the disruption matrices/vectors. (H) The lesion is segmented as contrast-enhancing (CE, red) and non-enhancing (NE, yellow) tumours. (I–K) The brain region disruptions are classified into CE, NE, distant and indirect disruptions. The WM connection disruption is classified into direct and indirect disruptions. (L) Categorical disrupted brain regions are mapped to the standard space. (M) Disrupted WM connections are mapped to the standard space producing a disruption density map.
Constructing a template of white matter connections
An unbiased WM connection template was generated by performing probabilistic tractography on the dMRI of the 10 healthy controls. In detail, the cortical/subcortical regions on dMRI were parcellated into 90 brain regions, using the Automatic Anatomical Labelling (AAL) atlas33 in the standard space.34 The AAL atlas included the grey-WM boundary to facilitate tractography. Deformable registration was performed using the Advanced Normalization Tools (ANTs).35
Eddy currents and subject motions in dMRI were corrected using the FSL eddy tool (v.6.0.0). A crossing fibre model was fitted to each control’s dMRI using the FSL function bedpostx. Probabilistic tractography between each pair of the 90 regions was subsequently performed using FSL Probtrackx2.36 Each region of interest (ROI) was used as a seed (starting ROI) and target (ending ROI) once in the tracking. For each pair of seed/target ROI, 5000 streamlines were sampled from the seed mask. Only the streamlines reaching the target mask were retained. The threshold of tracking curvature was set to 0.2 (80°). Streamline samples were terminated when they have travelled 2000 steps with step length of 0.5 mm or entered the cortical/subcortical regions. Streamlines were discarded if they entered the cerebrospinal fluids in the ventricle or re-entered the seed region.
Distribution maps were generated for all possible WM connections between the 90 cortical/subcortical regions in the healthy controls. The yielded distribution maps were non-linearly transformed to the MNI-152 space and averaged to mean connection distribution across the controls using fslmaths, which was thresholded to only retain the voxels with top 5% probability and finally binarized, providing conservative pathways for the template.
Generating skeletonized FA maps
In generating skeletonized FA maps, we included age-matched controls to reduce potential bias from ageing-related WM pathology. We fitted all dMRIs with a tensor model using the FSL diffusion toolbox.37 The yielded FA maps were non-linearly co-registered to a FA template in the standard space using the deformable function of ANTs, which could mitigate the brain deformation caused by tumour.38 This approach could outperform FNIRT39 (default registration tool in TBSS) in co-registering FA40 and pathology-bearing T1.41 To minimize potential the signal-noise ratio effect from different MRI protocols, we normalized the FA maps using the histogram-matching method.42
The local maxima voxels from the FA map of patients and controls were projected to this skeleton mask using an improved iterative TBSS projection approach guided by tract orientation.43 A FA skeleton mask in the standard space (FMRIB58 FA skeleton 1 mm) was used as the target for FA voxel projection.44 The generated individualized FA skeletons represent the centre integrity of WM tracts in subjects.
Estimating strengths of WM connections and brain regions
For each patient/control, the WM connection strength was calculated as the mean strength of all tract segments in the FA skeleton, constrained by the WM connection template. The columns and rows of each individualized connection matrix represent the brain regions, while the elements in the matrices (Cij) represent the strength of WM connection between the brain regions i and j. We calculated the strength Bi for region i, by aggregating the connectivity strength of the WM connection Cij connected to this region.
Identification of significantly disrupted connectome
We first calculated the mean strength and standard deviation (SD) of each WM connection across all healthy controls, which was compared to individual patients (Fig. 1G). We defined the significantly decreased connectome in patients as those with a strength of over 2SD (95% confidence) lower than the mean strength of the whole control group.
Estimating connectome disruptions
We calculated the patient-wise global disruption index by averaging the disruption of WM connections and brain regions, respectively. To address intra-tumour heterogeneity, we segmented the lesion as contrast-enhancing (CE, the entire area within the T1 CE rim) and peritumoral non-enhancing (NE, the hyper-intensities surrounding CE on FLAIR) subregions (Fig. 1H and I). Based on the segmented tumour, we categorized the disrupted WM connections and brain regions as follows (Fig. 1J and K):
Disrupted WM connections
Direct disruption: directly disrupted by tumour, travelling across the contrasting enhancing or NE tumour.
Indirect disruption: disrupted without crossing the lesion.
Disrupted brain regions
Tumour disrupted regions: disrupted regions within the lesion, further categorized into CE or NE disrupted regions, according to the overlap with tumour subregions.
Distant disrupted regions: disrupted regions within the normal-appearing brain and connected to the lesion via WM connections.
Indirect disrupted regions: disrupted regions without any connection to the lesion.
Finally, we calculated the regional disruption index as the average of each category.
Group distribution of tumour disruption
To generate a tumour frequency map, we non-linearly transformed all tumour masks from individual patients to the standard space using ANTs, with voxel-wise tumour distribution density normalized at the group level. Regional disrupted WM connections and brain regions are mapped to the standard space for visualization (Fig. 1L and M). We also quantified the disruption probability of brain regions and major tracts among patients. For tracts, we mapped the WM connections to the 42 anatomical tracts constructed from the 1000 healthy subjects available in the XTRACT toolbox of FSL. The disruption probability for each brain region or tract was calculated as the percentage of patients with this brain region or tract disrupted. We also calculated the proportion of disrupted connectomes out of all the patients.
Topological features of the connectome
We calculated two most commonly used topological features for each patient using the Brain Connectivity Toolbox17: characteristic path length and clustering coefficient. Briefly, the clustering coefficient measures the probability of two direct topological neighbours of a specific brain region being connected. The characteristic path length measures the average shortest path length of the network (see Supplementary material for formulas).45 We filtered the connectome with a population-consistency-based strength threshold to reduce the noise in feature calculation.46
Preserved connectivity of disrupted distant regions
By removing the disrupted WM connections, we investigated the preserved connections, defined as the remaining WM connections to the disrupted distant regions. Through pairwise comparison between patients and age-matched controls, we categorized patients into two subgroups with overall increased or decreased connectivity respectively, according to the summed strength of the preserved connectivities.
Statistical analysis
All analyses were performed in RStudio v.3.2.3 and MATLAB 2019b. The Benjamini–Hochberg procedure (BH–FDR) correction was performed for all multiple comparison tests. The hypothesis of no effect was rejected at a two-sided level of 0.05. The difference between global/regional disruptions and topological features of patient cohorts/subgroups were compared using two sample t-test. Pearson correlation was used to test the correlations between disruption indices, MMSE score, tumour volume and topological features. A multiple regression was used to identify significant regional disruptions contributing to topological changes. The mediation model was used to test whether the previously identified contributor mediated the effect of lesion on brain topology.
We performed survival analysis to evaluate disruption indices and topological features using OS and PFS. Patients alive at the last known follow-up were censored. Kaplan–Meier survival curves were compared using log-rank test, based on disruption indices and topological features dichotomized by either median or the optimal cut-off defined using the maximally selected rank statistics in R package ‘Survminer’,47 whichever was more significant.
In addition to the univariate Cox proportional hazards model, we conducted multivariate Cox proportional hazards model accounting for all relevant clinical covariates, including O-6-methylguanine-DNA methyltransferase (MGMT) methylation status, isocitrate dehydrogenase-1 (IDH-1) mutation, sex, age, the extent of resection, adjuvant therapy and tumour volume. We also included two tumour location features from the Visually Accessible Rembrandt Images (VASARI) feature set (https://wiki.cancerimagingarchive.net/display/Public/TCGA-GBM) describing the involvement of eloquent cortex and deep WM to account for the effects of tumour cortical/subcortical brain regions.
We used the receiver operating characteristic curves to evaluate the accuracy in predicting OS. We fit a generalized linear model and calculated the area under the receiver operating characteristic curve (AUC). The model included the significant disruption indices and topological features from the univariate Cox proportional hazards models.
Data availability
Enquiries or requests for the data should be directed to the corresponding author.
Results
Subject characteristics
For the discovery cohort, we recruited 136 patients for preoperative MRI scanning. After excluding 19 patients according to the trial exclusion criteria, we finally included 117 of 136 (86.0%) patients (mean age 59 years, range 22–75 years, 89 males) for analysis. Six patients (5.1%) were lost to follow-up. The median OS was 392 (range 34–1932) days. The median PFS was 275 (range 13–1393) days.
For the validation cohort, 49 patients were initially recruited and 42 patients were finally included according to the trial exclusion criteria (mean age 59 years, range 28–75 years, 30 males). Ten patients (23.8%) were alive or lost to follow-up. The median OS was 335 (range 55–962) days. The median PFS was 246 (range 21–805) days. Two study cohorts showed no significant differences in clinical variables (Supplementary Table 1).
We included 117 healthy age-matched subjects (mean age 59.9 years, 78 males) from the IXI datasets as healthy controls. In addition, we included 10 healthy subjects (mean age 60.9 years, 5 males) from the ADNI for the tractography template. All cohorts showed no significant difference in age.
Global disrupted connectome demonstrates clinical significance
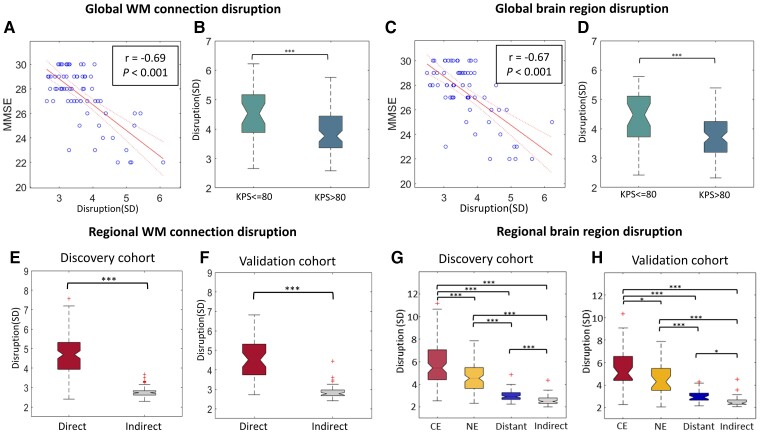
We observed that the global disruption of WM connections (Pearson: r = −0.67, P < 0.001; Spearman: rho = −0.48, P < 0.001; Fig. 2A) and brain regions (Pearson: r = −0.69, P < 0.001; Spearman: rho = −0.52, P < 0.001; Fig. 2C) were both negatively correlated with the MMSE score. To exclude the potential effect of age on cognition, a linear regression model was used to predict the MMSE score based on the disruption indices adjusting for age (MMSE ∼ disruption indices + Age). The results show that only the disruption indices were significant (Age: P = 0.296, Global WM disruption: P < 0.001; Age: P = 0.425, Global brain region disruption: P < 0.001). The correlation test between age and MMSE score showed no significance (Pearson: r = −0.17, P = 0.156; Spearman: rho = −0.17, P = 0.176).
Figure 2.
The clinical significance of disruption indices. (A and C) The global disruption indices of WM connections and brain regions are both negatively correlated with the MMSE score. (B and D) Higher disruption is associated with worse Karnofsky Performance Status. (E and F) In both discovery and validation cohorts, direct connection disruption is higher than Indirect disruption. (G and H) Disruption of tumour regions (CE, NE) are more significantly disrupted than the normal-appearing brain (distant, indirect). ***P < 0.001.
We further compared the patient subgroups stratified by the Karnofsky Performance Status (KPS) score of 80 as reported.48 We found that a worse KPS score was associated with higher disruptions of both brain regions and WM connections (both P < 0.001; Fig. 2B and D). We also found significant negative correlations between the KPS score and the global disruption indices (WM disruption: Pearson, r = −0.51, P < 0.001; Spearman: rho = −0.41, P < 0.001, Brain region disruption: Pearson, r = −0.49, P < 0.001; Spearman: rho = −0.31, P < 0.001). These results indicate that the stratified patient subgroups may have different global disruption indices.
Finally, we trained logistic regression models to predict the better or worse KPS as stratified previously. The baseline model including two preoperatively available variables: age and tumour volume, achieved an AUC of 0.79 [confidence interval (CI): 0.68–0.91]. Adding the global disruption indices into the model improved the AUC to 0.86 (CI: 0.77–0.94; Supplementary Fig. 1A). The out-of-sample prediction on the validation cohort showed improved performance (AUC = 0.71, CI: 0.53–0.89) compared to the baseline (AUC = 0.60, CI: 0.43–0.78; Supplementary Fig. 1B).
To rigorously assess the reproducibility of WM connection strength estimation, we compared the strengths of a cerebellar tract (middle cerebellar peduncle, MCP) that is not affected by the supratentorial tumours in our cohorts. No significant difference was found between controls and patients; Supplementary Fig. 2).
Regional disrupted connectome characteristics
Tumour regions are more significantly disrupted than distant regions
The two study cohorts showed no significant difference in disruption indices (Supplementary Table 2). In comparing disruptions of WM connections, we observed significantly higher direct disruption (4.77 ± 1.56) than indirect disruption (2.59 ± 0.39, P < 0.001; Fig. 2E, F and Supplementary Table 3). Similarly for brain regions, focal tumour (CE: 5.83 ± 2.00; NE: 4.67 ± 1.33) were more significantly disrupted than the normal-appearing brain (distant: 2.90 ± 0.71, indirect: 2.56 ± 0.39, each P <0.001; Fig. 2G and H). The validation cohort showed similar disruption patterns. These results correspond to our understanding of tumour invasion and support the robustness of the regional disruption indices.
The regional disruptions are correlated with focal tumour volume
Pearson correlation tests showed that the WM connection disruptions in tumour (direct) and normal-appearing brain (indirect) were positively correlated (r = 0.44, P < 0.001). Similarly, the disruption of distant regions was positively correlated with that of tumour regions (distant versus CE: r = 0.43, P < 0.001; distant versus NE: r = 0.34, P = 0.028; Supplementary Table 4). Further, the tumour volume (measured by CE tumour) was positively correlated with the disruptions of both direct connections (r = 0.52, P < 0.001) and distant regions (r = 0.33, P < 0.001; Supplementary Table 5). Collectively, these data indicate that a larger focal tumour is associated with higher connectome disruption throughout the brain.
The normal-appearing brain shows widespread disruption
We calculated the proportion of disrupted regions out of all brain regions. It is worth noting that, in the group analysis, a higher proportion of distant regions (16.8 ± 12.0%) was identified than focal lesion (CE: 5.8 ± 5.1%, P < 0.001), recapitulated by the validation cohort (Supplementary Table 6), indicating that the normal-appearing brain was widely disrupted.
At the group level, the average disruption probability of distant regions was higher (17.2 ± 9.0%) than focal lesion (CE: 11.8 ± 6.8%, P < 0.001), possibly due to the more extensive coverage of the distant regions. This finding further confirmed that the disruption of brain regions was widespread beyond the lesion.
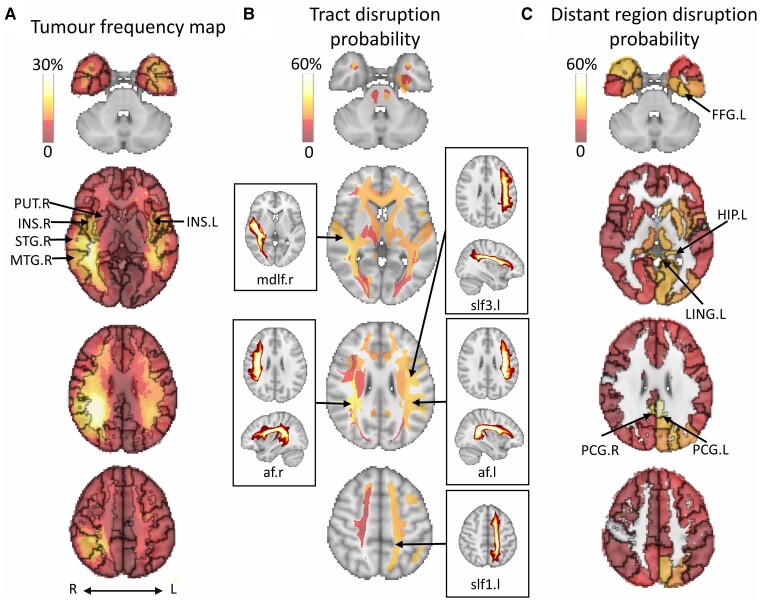
We further compared the tumour frequency map (Fig. 3A) with the distribution probability map of brain regions and tracts. Notably, the top five tracts most likely disrupted were mainly association tracts and close to the high-frequency regions (Fig. 3B and Supplementary Table 7), suggesting that the association tracts may be more vulnerable in glioblastoma patients. The top five most probably disrupted distant regions (Fig. 3C) were mainly in the low-frequency regions (see Supplementary Table 8 for details).
Figure 3.
Tumour density and disrupted anatomical structures. (A) Tumour frequency maps are generated using tumour segmentation masks. The top five disrupted focal brain regions include the right superior temporal gyrus (STG.R), right middle temporal gyrus (MTG.R), right insula (INS.R), left insula (INS.L) and right lenticular nucleus, putamen (PUT.R). (B) The top five disrupted anatomical tracts and their maximum intensity projection: right arcuate fasciculus (af.r), right middle longitudinal fasciculus (mdlf.r), left superior longitudinal fasciculus 3 (slf3.l), left arcuate fasciculus (af.l) and left superior longitudinal fasciculus 1 (slf1.l). (C) The top five disrupted distant regions include the left posterior cingulate gyrus (PCG.L), right posterior cingulate gyrus (PCG.R), left lingual gyrus (LING.L), left fusiform gyrus (FFG.L) and left hippocampus. L/l = left; R/r = right. ***P < 0.001, **P < 0.01, *P < 0.05.
Topological features and their association with distant regions
The focal tumour alters the topological property of the connectome
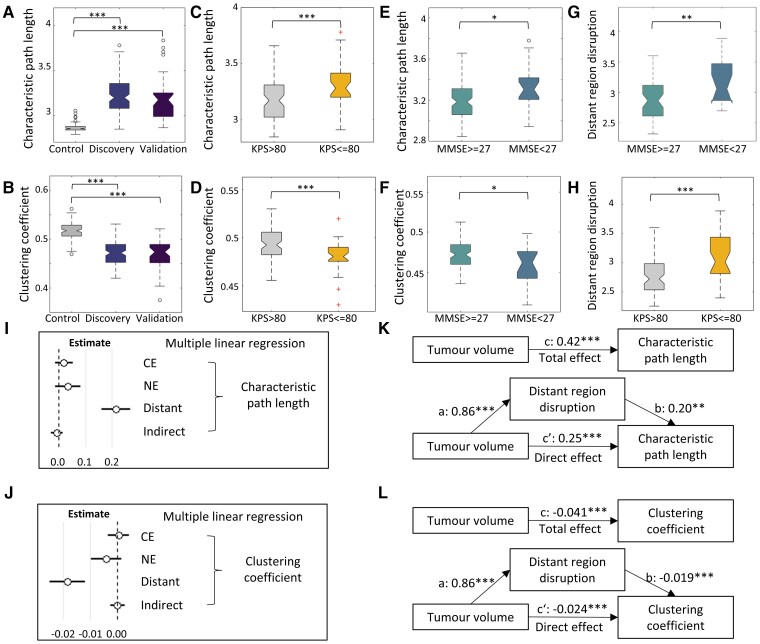
We observed significantly higher characteristic path lengths in patient networks than healthy controls (P < 0.001; Fig. 4A). In contrast, the clustering coefficient of patients was significantly lower than that of healthy controls (P < 0.001; Fig. 4B and Supplementary Table 9). These results reveal that tumour lesions could dramatically alter the topology property of the structural connectome. No difference was found between topological features between two patient cohorts, which suggests the robustness of our approach.
Figure 4.
Topological alteration of the connectome. Patients show increased characteristic path length (A) and decreased clustering coefficient (B) compared to the controls. Patient subgroups with worse preoperative KPS (C and D) and MMSE (E and F) scores show increased characteristic path length and decreased clustering coefficient. Disruption of distant regions is higher in the subgroups with worse MMSE (G) and KPS (H), and it is the only significant predictor of characteristic path length (I) and clustering coefficient (J) in multiple linear regression. (K) The effects of tumour volume on characteristic path length are mediated by the disruption of distant regions: total effect (c path) = 0.42, P < 0.001; direct effect (c′ path) = 0.25, P < 0.001; mediation effect (c—c′) = 0.17, P = 0.008. (L) The effects of tumour volume on clustering coefficient are mediated by the disruption of distant regions: total effect (c path) = −0.041, P < 0.001; direct effect (c′ path) = −0.024, P < 0.001; mediation effect (c—c′) = −0.017, P < 0.001. ***P < 0.001, **P < 0.01, *P < 0.05.
We next determined the clinical significance of connectome topology by comparing the topological properties of the patient subgroups stratified by MMSE and KPS scores. We found that the patients with lower MMSE or KPS scores presented lower clustering coefficient (MMSE: P = 0.012, KPS: P < 0.001; Fig. 4C and E) and higher characteristic path length (MMSE: P = 0.013, KPS: P < 0.001; Fig. 4D and F). Moreover, characteristic path length (r = 0.43, P < 0.001) was positively correlated with tumour volume, while clustering coefficient (r = −0.45, P < 0.001) was negatively correlated with tumour volume, indicating that a larger focal lesion may have a greater influence on the connectome topology (Supplementary Table 10).
The disruption of distant regions is associated with the topological alteration
To understand the relation between the regional disruption with topological properties, we performed a multiple linear regression, which revealed that the disruption of distant regions was the only significant predictor of characteristic path length (estimate = 0.21, P < 0.001) and clustering coefficient (estimate = −0.018, P < 0.001; Fig. 4I and J).
We further performed the mediation analysis, which showed that tumour volume had both significant direct and indirect effects (mediated by the disruption of distant regions) on characteristic path length (direct: P < 0.001, indirect P = 0.008) and clustering coefficient (direct and indirect P < 0.001) (Fig. 4K and L). The findings were confirmed by the validation cohort (Supplementary Fig. 3).
We additionally compared the disruptions of distant regions in the subgroups stratified by the MMSE and KPS scores. We noticed that the patients with higher MMSE or higher KPS scores displayed significantly lower disruption of distant regions (Fig. 4G and H), consistent with the distinct network topological properties in these subgroups. The results further indicate the association between the disrupted distant regions and connectome topology.
Topological features and disruption of distant regions are prognostic
For both disruption indices and topological features, we evaluated the prognostic value using log-rank tests and Cox proportional hazards models.
Log-rank test
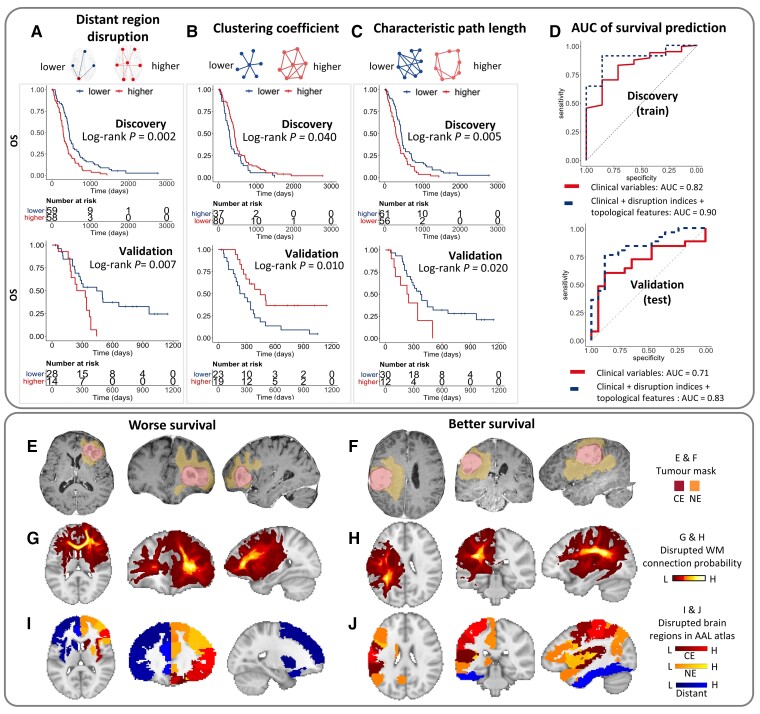
In the subgroups stratified by the mean disruption of distant regions (2.9), patients with higher disruption had worse survival than lower disruption (OS: median 293 versus 449 days, P = 0.002, PFS: median 238 versus 307 days, P = 0.019; Fig. 5A). Further, the subgroups stratified by the optimal cut-off of topological features (clustering coefficient 0.46; characteristic path length 3.20) also had distinct survival. Precisely, the subgroup with a higher clustering coefficient had better survival than that with a lower clustering coefficient (OS: median 475 versus 294 days, P = 0.040, PFS: median 306 versus 238 days, P = 0.002; Fig. 5B). The subgroup with lower characteristic path length showed better survival than that with higher characteristic path length (OS: median 465 versus 288 days, P = 0.005, PFS: median 312 versus 244 days, P = 0.012; Fig. 5C). We further confirmed the findings in the validation cohort using identical cut-offs.
Figure 5.
The prognostic value of disruption indices and topological features. The top shows that in both cohorts, higher disruption of distant regions (A), lower clustering coefficient (B) and higher characteristic path length (C) are associated with worse OS. (D) The model of predicting OS using clinical factors, disruption indices and topological features show improved AUC compared to clinical factors alone. Bottom shows two examples with worse or better survival (OS: 317 versus 1555 days; PFS: 159 versus 747 days). Both are IDH wild-type and MGMT unmethylated tumours of similar visible size in two males (aged 67 versus 69 years), who underwent complete resection followed by temozolomide chemoradiotherapy (E and F). Patients have similar tumour size on T1C (23.6 versus 25.0 cm3). The patient with worse survival (G) has more widespread connection disruption beyond the visible lesion, compared to the patient with better survival (H). The disruption indices of distant regions (blue) are 3.0 (I) and 2.8 (J), respectively. Their topological features are distinct (clustering coefficient 0.44 versus 0.48: characteristic path length 3.31 versus 3.17). HR = hazard ratio.
Cox proportional hazards modelling
From the Cox proportional hazards models (Table 1), we observed that higher disruptions of indirect connection (OS: HR = 1.36, P = 0.007; PFS: HR = 2.43, P = 0.046) and distant regions (OS: HR = 1.46, P = 0.049; PFS: HR = 1.49, P = 0.019) were associated with worse survival. For topological features, a higher clustering coefficient was associated with better survival (OS: HR = 0.63, P = 0.035; PFS: HR = 0.49, P = 0.002), while a higher characteristic path length was associated with worse survival (OS: HR = 1.56, P = 0.035; PFS: HR = 1.82, P = 0.009).
Table 1.
Univariate survival statistics of the discovery cohort
| OS | PFS | |||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P | HR | 95%CI | P | |
| Clinical variables | ||||||
| Age | 1.03 | 1.01 –1.05 | 0.004 | 1.03 | 1.00–1.05 | 0.021 |
| Sexa | 0.81 | 0.52–1.25 | 0.333 | 0.75 | 0.47–1.19 | 0.217 |
| Performanceb | 1.60 | 1.09–2.36 | 0.018 | 1.63 | 1.04–2.55 | 0.033 |
| IDHc | 0.59 | 0.25–1.36 | 0.211 | 0.52 | 0.22–1.21 | 0.131 |
| MGMTd | 0.77 | 0.52–1.14 | 0.196 | 0.68 | 0.43–1.07 | 0.094 |
| EORe | 1.89 | 1.26–2.84 | 0.002 | 1.90 | 1.19–3.03 | 0.007 |
| Adjuvant treatmentf | 0.21 | 0.13–0.34 | <0.001 | 0.22 | 0.11–0.41 | <0.001 |
| Tumour volume | 1.01 | 1.00–1.01 | 0.001 | 1.01 | 1.00–1.02 | 0.045 |
| Eloquent locationg | 0.93 | 0.64–1.36 | 0.711 | 1.06 | 0.69–1.62 | 0.807 |
| Deep WMh | 0.85 | 0.58–1.24 | 0.386 | 0.86 | 0.57–1.31 | 0.487 |
| NE volume | 0.87 | 0.72–1.05 | 0.135 | 0.87 | 0.70–1.07 | 0.192 |
| Disruption indices | ||||||
| Direct connection | 1.07 | 0.87–1.30 | 0.790 | 1.08 | 0.88–1.32 | 0.464 |
| Indirect connection | 1.36 | 1.13–1.65 | 0.007 | 2.43 | 1.02–5.81 | 0.046 |
| CE regions | 0.95 | 0.86–1.04 | 0.667 | 0.96 | 0.86–1.06 | 0.418 |
| NE regions | 1.01 | 0.88–1.15 | 0.939 | 1.00 | 0.87–1.15 | 0.979 |
| Distant regions | 1.46 | 1.08–1.99 | 0.049 | 1.49 | 1.07–2.07 | 0.019 |
| Indirect regions | 1.06 | 0.87–1.29 | 0.790 | 0.80 | 0.83–1.28 | 0.795 |
| Topological features | ||||||
| Clustering coefficient | 0.63 | 0.42–0.93 | 0.035 | 0.49 | 0.30–0.78 | 0.002 |
| Characteristic path length | 1.56 | 1.06–2.29 | 0.035 | 1.82 | 1.16–2.84 | 0.009 |
EOR = extent of resection. Bold values = P < 0.05.
Female as the reference.
KPS 90–100 as the reference.
IDH wild-type as reference.
Unmethylated MGMT as reference.
Incomplete resection as reference.
Concurrent chemoradiotherapy as reference.
Non-eloquent location as reference.
Affected deep WM as reference.
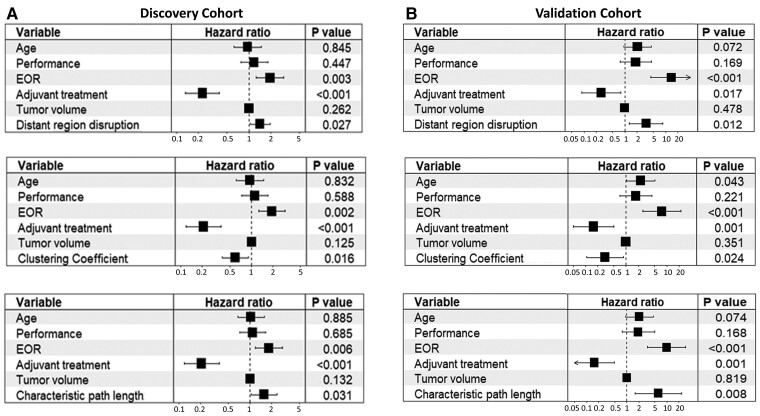
In the multivariate model adjusting for all the significant clinical covariates from the univariate models, the disruption of distant regions and topological features remained significant (Fig. 6 and Supplementary Table 11). Their prognostic value was confirmed by the validation cohort (Supplementary Tables 12 and 13).
Figure 6.
Forest plots of multivariate survival modelling. For the discovery (A) and validation (B) cohorts, the higher disruption of distant regions, higher characteristic path length and lower clustering coefficient are associated with worse survival. The prognostic value is independent of the significant clinical variables. EOR = extent of resection.
Logistic model for survival prediction
We trained a logistic regression model on the discovery cohort to predict the 2-year OS using the disruption indices and topological features significant in the univariate Cox model. The baseline model, including the previous significant clinical variables (i.e. age, extent of resection and adjuvant therapy), achieved an AUC of 0.82 (CI 0.68–0.96). By adding the disruption indices and topological features into the baseline model, the AUC was improved to 0.90 (CI 0.80–0.99; Fig. 5D). We further tested this trained model in the validation cohort for out-of-sample validation, which confirmed the improved accuracy (AUC = 0.83, CI 0.71–0.96) by including disruption indices and topological features over the baseline model (AUC = 0.71, CI 0.54–0.87).
We presented two examples (Fig. 5E–J) with similar clinical variables but different disruption of distant regions, topological features and finally distinct survival (above and below the median, respectively).
Preserved connectivity and recurrence distance associated with distant regions
The preserved connectome of distant regions indicates patient survival
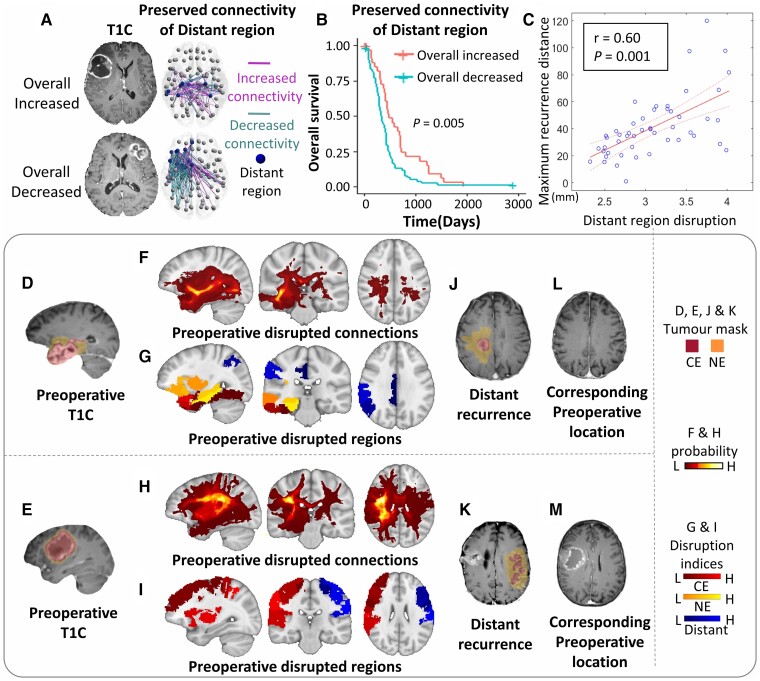
As the distant region disruption is the most significant biomarker, we further analysed its potential effect on the preserved connectome. First, we observed that 93.2% (109/117) patients displayed overall changes in connectivity. Among them, 24.7% (29/117) patients displayed overall increased connectivity, while 68.4% (80/117) patients showed overall decreased connectivity. We present two case examples with overall increased and decreased connectivity in the preserved connectome of the distant regions, respectively (Fig. 7A). The log-rank test showed that those patients with overall increased connectivity were associated with better survival (P = 0.005; Fig. 7B), confirmed by the validation cohort (Supplementary Fig. 4). The findings suggest that more integrated brain connectivity is associated with better patient survival, suggesting a priori more resilient connectome or network reorganization.
Figure 7.
The disruptions of the distant regions indicate survival significance and distant recurrence. The top shows that after removing the disrupted WM connections of the distant regions, the preserved connections are categorized as increased or decreased connectivity in comparisons with healthy controls and aggregated to stratify patients. Two examples of overall increased and decreased connectivity are in A. The subgroup with overall increased connectivity shows better survival than overall decreased connectivity (B). The disruption index of the distant region is positively correlated with maximum recurrence distance (C). The bottom shows two examples of distant recurrence. Both patients present solitary visible lesions on preoperative T1C images (D and E), widespread disrupted WM connections (F and H) and brain regions (G and I). In both patients, the distant recurrence location, either ipsilesional recurrence (J) or contralesional recurrence (K), corresponds to the distant regions (blue), which are linked to the primary lesion via the WM connections shown in F and H. A retrospective review of the preoperative T1C images reveals no visible lesion in the recurrence location (L and M). T1C = T1 contrast.
Disrupted connectome indicates tumour recurrence
Finally, we found that the higher distant region disruption was positively correlated with the furthest recurrence distance from tumour centroid (r = 0.60, P < 0.001; Fig. 7C). We present two cases that showed distant recurrence in follow-up scans, where the disrupted distant regions indicated occult tumour invasion invisible on the preoperative MRI (Fig. 7D–M).
Discussion
The present study employed a connectome approach to investigate the disruption of structural connectivity in glioblastoma. Our main findings include: (i) glioblastomas cause widespread disrupted neural connectivity beyond the focal lesion; (ii) the disruption of the normal-appearing brain could mediate the alteration of connectome topology, associated with worse patient performance and affect patient survival; and (iii) the preserved connectome demonstrates evidence of network reorganization associated with survival.
The finding that glioblastomas can cause widespread structural impairment is in line with the previous studies using resting-state fMRI, reporting that glioma induced widespread functional impairment.3,4 The evidence supports that glioblastoma should be treated as a systematic disease rather than a local disease. Moreover, we found that only the disruption of distant regions was associated with topological alteration and patient survival among all the regional disruptions, suggesting the importance of characterizing global neural connectivity.
In the anatomical mapping of the disrupted connectome, we found that the top disrupted distant regions, e.g. posterior cingulate cortex and hippocampus, are essential structures of the limbic system, suggesting the propensity of the occult invasion affecting the limbic system. Moreover, the top affected anatomical tracts, e.g. arcuate fasciculus and superior longitudinal fasciculus, are long association tracts widely connecting separated gyri, suggesting that tumour invasion might spread through these tracts. Although at the macroscopic scale, our imaging findings may provide a perspective for previously reported neural–cancer interaction.49
We found that the connectivity measures could provide superior biomarkers for brain tumour stratification over conventional clinical factors, e.g. tumour location and volume. The network efficiency of the human brain generally reflects the integrity of brain function.50 Glioblastoma patients displayed decreased network efficiency compared to healthy controls, probably due to tumour disturbance on brain function. Interestingly, our results show that the preserved connectome demonstrates evidence of reorganization. The increased connectivity, indicating a more integrated network and more robust function, is associated with favourable survival. Although the mechanism remains further elucidated, it could suggest the opportunities of understanding neural–cancer interaction for patient prognosis.
Our study has important clinical implications. Due to the remarkable heterogeneity of glioblastoma, the development of quantitative prognostic markers is crucial for precise diagnosis and treatment. The structural connectome and topological features confer a novel approach to investigate the systematic changes of neural connectivity in glioblastoma. It could enable us to understand the interaction between tumour invasion and neural connectivity, which promises to stratify patients more precisely, help to develop targeted therapeutics and reduce neurological deficits. From a translational perspective, our approach shows the potential to quickly produce metrics to enhance prognosis determination and patient stratification through automated imaging analytical software in the clinic, incorporating various data processing and modelling procedures, which could assist clinical decision making in future. Additionally, tumour invasion in the normal-appearing brain visualized on the imaging could be integrated into the neuro-navigation and radiotherapy planning systems, offering more precise guidance for treatment targets.
Our study has limitations. First, structural connectome can only directly measure the connectivity of connecting tracts. Although most brain regions are connected via tracts, some functionally related regions may not be structurally connected. Future work could include resting-state MRI and functional connectivity. Second, we only included de novo glioblastoma who received first-line treatment in the trial. Molecular markers, i.e. IDH and MGMT methylation, were not significant as previously reported in our cohorts. Finally, neuronal degeneration, such as Wallerian Degeneration, may also lead to the disruptions of WM connections, which may not be differentiated by our approach. Nonetheless, the significant prognostic value of disruption indices could support the capability of our approach in indicating tumour invasion.
In conclusion, glioblastoma causes widespread impairment to the structural connectome. The invisible disruption on conventional MRI and connectome integrity are correlated with patient survival. Studying neural connectivity may provide a valuable tool for patient stratification and precise treatment for patients with brain tumours.
Supplementary Material
Acknowledgements
Part of the data were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing. The ADNI was launched in 2003 as a public-private partnership led by Principal Investigator Michael W. Weiner, MD. A complete listing of investigators can be found at http://adni.loni.usc.edu/. For up-to-date information, see www.adni-info.org. Data collection and sharing were funded by ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Contributor Information
Yiran Wei, Cambridge Brain Tumour Imaging Laboratory, Division of Neurosurgery, Department of Clinical Neurosciences, University of Cambridge, Cambridge CB2 0QQ, UK.
Chao Li, Cambridge Brain Tumour Imaging Laboratory, Division of Neurosurgery, Department of Clinical Neurosciences, University of Cambridge, Cambridge CB2 0QQ, UK; Department of Applied Mathematics and Theoretical Physics, The Centre for Mathematical Imaging in Healthcare, Cambridge CB3 0WA, UK.
Zaixu Cui, Chinese Institute for Brain Research, Beijing 102206, China.
Roxanne Claudeve Mayrand, Cambridge Brain Tumour Imaging Laboratory, Division of Neurosurgery, Department of Clinical Neurosciences, University of Cambridge, Cambridge CB2 0QQ, UK.
Jingjing Zou, Division of Biostatistics and Bioinformatics, Department of Family Medicine and Public Health, University of California, San Diego, CA 92103, USA.
Adrianna Leanne Kok Chi Wong, Cambridge Brain Tumour Imaging Laboratory, Division of Neurosurgery, Department of Clinical Neurosciences, University of Cambridge, Cambridge CB2 0QQ, UK.
Rohitashwa Sinha, Cambridge Brain Tumour Imaging Laboratory, Division of Neurosurgery, Department of Clinical Neurosciences, University of Cambridge, Cambridge CB2 0QQ, UK.
Tomasz Matys, Department of Radiology, University of Cambridge, Cambridge CB2 0QQ, UK.
Carola-Bibiane Schönlieb, Department of Applied Mathematics and Theoretical Physics, The Centre for Mathematical Imaging in Healthcare, Cambridge CB3 0WA, UK.
Stephen John Price, Cambridge Brain Tumour Imaging Laboratory, Division of Neurosurgery, Department of Clinical Neurosciences, University of Cambridge, Cambridge CB2 0QQ, UK.
Funding
This study was supported by the National Institute for Health Research (NIHR) (Brain Injury MedTech Co-operative). S.J.P. acknowledges National Institute for Health Research (NIHR) (Career Development Fellowship, CDF-18-11-ST2-003). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. C.L. acknowledges the Guarantors of Brain Fellowship. C.B.S. acknowledges Engineering and Physical Sciences Research Council (EP/M00483X/1 and EP/N014588/1). R.S. acknowledges Cancer Research UK (RRZB/040) and the Royal College of Surgeons of England (RRAG/093).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre J-Y. Primary brain tumours in adults. Lancet. 2012;379:1984–1996. [DOI] [PubMed] [Google Scholar]
- 2. Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daniel AG, Park KY, Roland JL, et al. Functional connectivity within glioblastoma impacts overall survival. Neuro Oncol. 2020;23:412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stoecklein VM, Stoecklein S, Galiè F, et al. Resting-state fMRI detects alterations in whole brain connectivity related to tumor biology in glioma patients. Neuro Oncol. 2020;22:1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aerts H, Fias W, Caeyenberghs K, Marinazzo D. Brain networks under attack: Robustness properties and the impact of lesions. Brain. 2016;139:3063–3083. [DOI] [PubMed] [Google Scholar]
- 6. Yu K, Lin CJ, Hatcher A, et al. PIK3CA Variants selectively initiate brain hyperactivity during gliomagenesis. Nature. 2020;578:166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Venkataramani V, Tanev DI, Strahle C, et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature. 2019;573:532–538. [DOI] [PubMed] [Google Scholar]
- 8. Price S, Burnet N, Donovan T, et al. Diffusion tensor imaging of brain tumours at 3 T: A potential tool for assessing white matter tract invasion? Clin Radiol. 2003;58:455–462. [DOI] [PubMed] [Google Scholar]
- 9. Price SJ, Pena A, Burnet NG, et al. Tissue signature characterisation of diffusion tensor abnormalities in cerebral gliomas. Eur Radiol. 2004;14:1909–1917. [DOI] [PubMed] [Google Scholar]
- 10. Li C, Wang S, Yan JL, et al. Characterizing tumor invasiveness of glioblastoma using multiparametric magnetic resonance imaging. J Neurosurg. 2019;132:1465–1472. [DOI] [PubMed] [Google Scholar]
- 11. Li C, Yan JL, Torheim T, et al. Low perfusion compartments in glioblastoma quantified by advanced magnetic resonance imaging and correlated with patient survival. Radiother Oncol. 2019;134:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li C, Wang S, Yan JL, et al. Intratumoral heterogeneity of glioblastoma infiltration revealed by joint histogram analysis of diffusion tensor imaging. Neurosurgery. 2019;85:524–534. [DOI] [PubMed] [Google Scholar]
- 13. Yan JL, Li C, van der Hoorn A, Boonzaier NR, Matys T, Price SJ. A neural network approach to identify the peritumoral invasive areas in glioblastoma patients by using MR radiomics. Sci Rep. 2020;10:9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Dijken BRJ, Yan JL, Boonzaier NR, et al. Subventricular zone involvement characterized by diffusion tensor imaging in glioblastoma. World Neurosurg. 2017;105:697–701. [DOI] [PubMed] [Google Scholar]
- 15. Price SJ, Allinson K, Liu H, et al. Less invasive phenotype found in isocitrate dehydrogenase-mutated glioblastomas than in isocitrate dehydrogenase wild-type glioblastomas: A diffusion-tensor imaging study. Radiology. 2017;283:215–221. [DOI] [PubMed] [Google Scholar]
- 16. Bullmore E, Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. [DOI] [PubMed] [Google Scholar]
- 17. Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage. 2010;52:1059–1069. [DOI] [PubMed] [Google Scholar]
- 18. Crossley NA, Mechelli A, Scott J, et al. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137:2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raj A, LoCastro E, Kuceyeski A, et al. Network diffusion model of progression predicts longitudinal patterns of atrophy and metabolism in Alzheimer’s disease. Cell Rep. 2015;10:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun Y, Chen Y, Collinson SL, Bezerianos A, Sim K. Reduced hemispheric asymmetry of brain anatomical networks is linked to schizophrenia: A connectome study. Cereb Cortex. 2017;27:602–615. [DOI] [PubMed] [Google Scholar]
- 21. Liu Y, Yang K, Hu X, et al. Altered rich-club organization and regional topology are associated with cognitive decline in patients with frontal and temporal gliomas. Front Hum Neurosci. 2020;14:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aerts H, Schirner M, Dhollander T, et al. Modeling brain dynamics after tumor resection using The Virtual Brain. Neuroimage. 2020;213:116738. [DOI] [PubMed] [Google Scholar]
- 23. Kinno R, Ohta S, Muragaki Y, Maruyama T, Sakai KL. Differential reorganization of three syntax-related networks induced by a left frontal glioma. Brain. 2014;137:1193–1212. [DOI] [PubMed] [Google Scholar]
- 24. Mandal AS, Romero-Garcia R, Hart MG, Suckling J. Genetic, cellular, and connectomic characterization of the brain regions commonly plagued by glioma. Brain. 2020;143:3294–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu L, Zhang H, Rekik I, Chen X, Wang Q, Shen D. Outcome prediction for patient with high-grade gliomas from brain functional and structural networks. Med Image Comput Comput Assist Interv. 2016;9901:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gorlia T, van den Bent MJ, Hegi ME, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: Prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE. 3. Lancet Oncol. 2008;9:29–38. [DOI] [PubMed] [Google Scholar]
- 27. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. [DOI] [PubMed] [Google Scholar]
- 28. Kamnitsas K, Ferrante E, Parisot S, et al. DeepMedic for brain tumor segmentation. In: Crimi A and Bakas S, eds. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Springer; 2016:138–149. [Google Scholar]
- 29. Schult T, Hauser T-K, Klose U, Hurth H, Ehricke H-H. Fiber visualization for preoperative glioma assessment: Tractography versus local connectivity mapping. PLoS ONE. 2019;14:e0226153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith SM, Jenkinson M, Johansen-Berg H, et al. Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. [DOI] [PubMed] [Google Scholar]
- 31. Fagerholm ED, Hellyer PJ, Scott G, Leech R, Sharp DJ. Disconnection of network hubs and cognitive impairment after traumatic brain injury. Brain. 2015;138:1696–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Squarcina L, Bertoldo A, Ham TE, Heckemann R, Sharp DJ. A robust method for investigating thalamic white matter tracts after traumatic brain injury. Neuroimage. 2012;63:779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. [DOI] [PubMed] [Google Scholar]
- 34. Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL. Symmetric atlasing and model based segmentation: An application to the hippocampus in older adults. Med Image Comput Comput Assist Interv. 2006;9(Pt 2):58–66. [DOI] [PubMed] [Google Scholar]
- 35. Avants BB, Tustison N, Song G. Advanced normalization tools (ANTS). Insight J. 2009;2:1–35. [Google Scholar]
- 36. Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prasanna P, Mitra J, Beig N, et al. Mass Effect Deformation Heterogeneity (MEDH) on gadolinium-contrast T1-weighted MRI is associated with decreased survival in patients with right cerebral hemisphere glioblastoma: A feasibility study. Sci Rep. 2019;9:1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andersson JL, Jenkinson M, Smith S. Non-linear registration aka Spatial normalisation FMRIB Technical Report TR07JA2. FMRIB Analysis Group of the University of Oxford. 2007:1–22.
- 40. Schwarz CG, Reid RI, Gunter JL, et al. Improved DTI registration allows voxel-based analysis that outperforms tract-based spatial statistics. Neuroimage. 2014;94:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ou Y, Akbari H, Bilello M, Da X, Davatzikos C. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: results and insights. IEEE Trans Med Imaging. 2014;33:2039–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Medical Imaging. 2000;19:143–150. [DOI] [PubMed] [Google Scholar]
- 43. Wei Y, Li C, Price SJ. Quantifying structural connectivity in brain tumor patients. In: Shen D, Liu T, Peters TM, et al., eds. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Springer; 2021:519–529. [Google Scholar]
- 44. Jenkinson M, Beckmann CF, Behrens T, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. [DOI] [PubMed] [Google Scholar]
- 45. Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. [DOI] [PubMed] [Google Scholar]
- 46. Roberts JA, Perry A, Roberts G, Mitchell PB, Breakspear M. Consistency-based thresholding of the human connectome. Neuroimage. 2017;145:118–129. [DOI] [PubMed] [Google Scholar]
- 47. Lin H, Zelterman D. Modeling survival data: extending the cox model. Taylor & Francis; 2002. [Google Scholar]
- 48. Chaichana K, Parker S, Olivi A, Quiñones-Hinojosa A. A proposed classification system that projects outcomes based on preoperative variables for adult patients with glioblastoma multiforme. J Neurosurg. 2010;112:997–1004. [DOI] [PubMed] [Google Scholar]
- 49. Monje M, Borniger JC, D’Silva NJ, et al. Roadmap for the emerging field of cancer neuroscience. Cell. 2020; 181:219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sporns O, Zwi JD. The small world of the cerebral cortex. Neuroinformatics. 2004;2:145–162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Enquiries or requests for the data should be directed to the corresponding author.