Fig. 4 |. Biochemical characterization of RG-I:GalAT activity by RGGAT1.
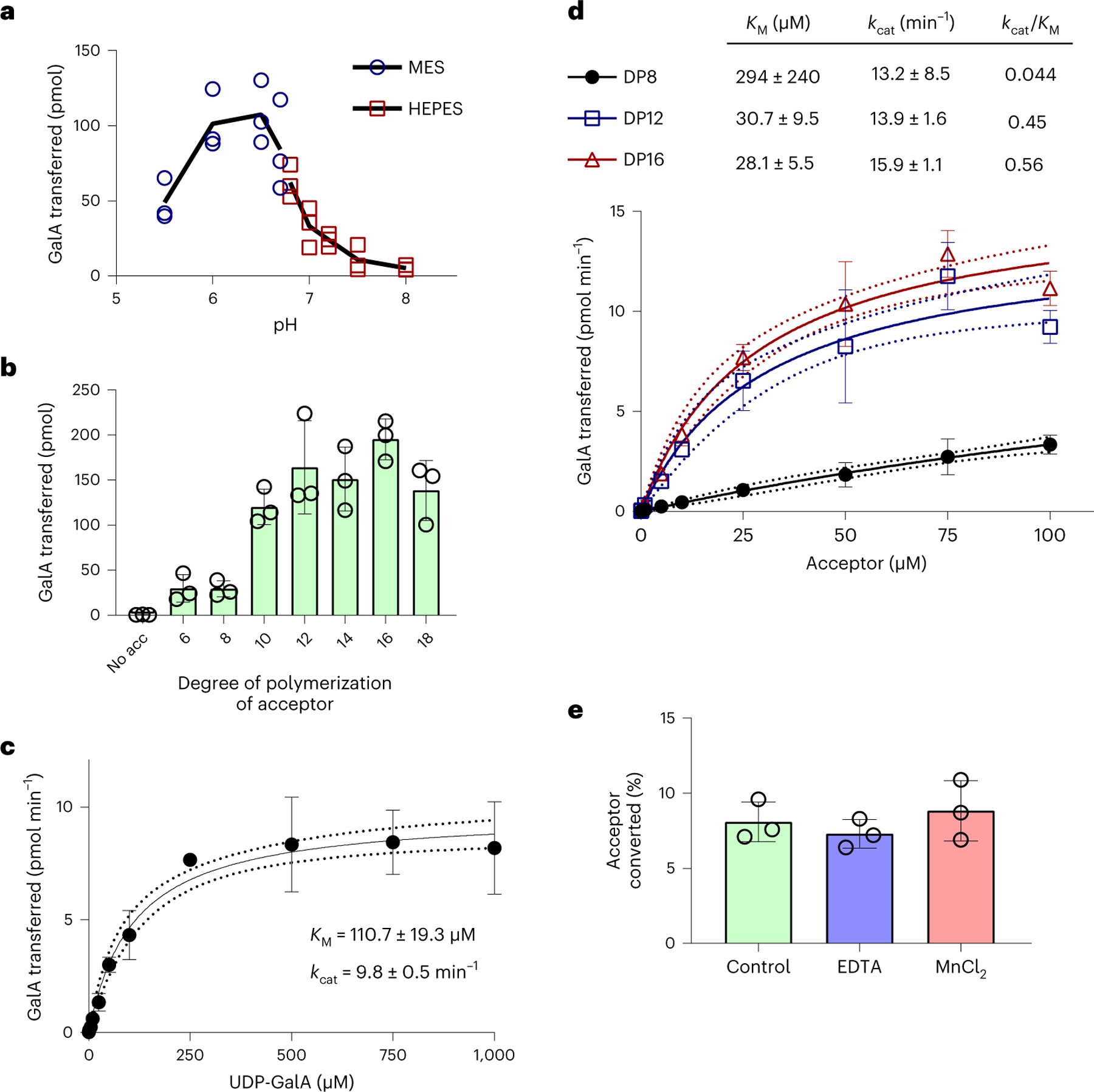
a, The pH optimum of RGGAT1 activity was measured using UDP-Glo in 10 min reactions containing 50 nM enzyme, 1 mM UDP-GalA and 100 µM of a DP12-2AB acceptor. Reactions were incubated with 50 mM of MES buffer (blue circles) of pH 5.5–6.7 or HEPES buffer (red squares) of pH 6.7–8.0. The buffer MES pH 6.5 was used for standard condition assays. The black line represents the average value of n = 3 independent assays. Individual data points from the three assays are shown. b, RGGAT1 activity was measured in 10 min reactions containing 50 nM enzyme, 1 mM UDP-GalA and 100 µM of acceptors with degrees of polymerization ranging from DP6 to DP18 or no acceptor (no acc) using UDP-Glo. Error bars represent standard deviations of n = 3 independent experiments. c, Michaelis-Menten kinetics for the UDP-GalA donor. RGGAT1 was incubated for 10 min with 100 µM DP16 acceptor and variable concentrations of UDP-GalA (0–1,000 µM). Kinetic constants were calculated by nonlinear regression using GraphPad Prism. Error bars represent standard deviations from n = 4 independent experiments. Dotted lines represent 95% confidence intervals. KM and kcat are reported as mean ± s.e.m. d, Michaelis-Menten kinetics for RG-I oligosaccharide acceptors of DP8, DP12 and DP16. RGGAT1 was incubated for 10 min with 1 mM UDP-GalA and variable concentrations of the indicated acceptors (0–100 µM). Kinetic constants were calculated by nonlinear regression using GraphPad Prism. Error bars represent standard deviations of n = 3 independent experiments. Dotted lines represent 95% confidence intervals. KM and kcat are reported as mean ± s.e.m. e, RGGAT1 was incubated in a 50 mM MES pH 6.5 buffer (control) or with buffer containing either 10 mM EDTA or 10 mM MnCl2 for 30 min before the assay. After a 30 min incubation period, the enzyme was diluted and assayed as described. The final concentration during the reaction was 10 mM for EDTA and 0.25 mM for MnCl2. No difference in activity compared to the control reaction was detected. Error bars represent standard deviations of n = 3 independent experiments.
