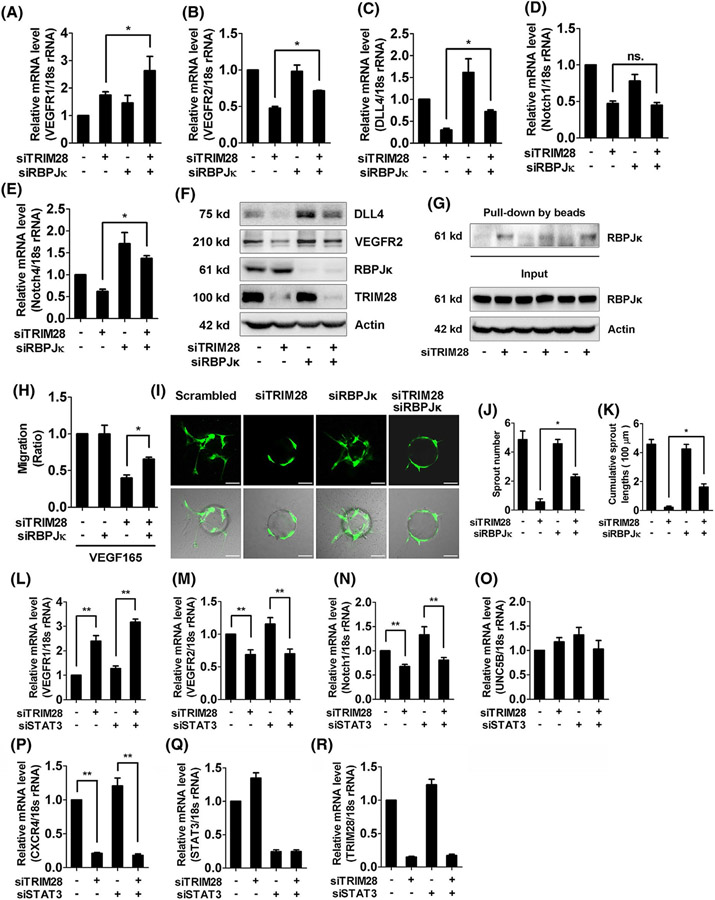
FIGURE 5.
TRIM28 regulates the interaction between HIF-1α and RBPJκ in ECs. A-F, ECs were transfected with the different siRNA fragments as indicated and harvested at 48 hours, and mRNA expressions of VEGFR1 (A), VEGFR2 (B), DLL4 (C), Notch1 (D), and Notch4 (E) were subjected to qRT-PCR. n = 3. *P < .05, ns., no significance. The protein expressions of DLL4, VEGFR2, RBPJκ, and TRIM28 were examined using western blot (F). n = 3. The representative images were shown. G, Nuclear extracts from control or TRIM28 knockdown ECs were incubated with oligonucleotides containing HIF-1α-binding site. The precipitations were checked using western blot with a RBPJκ antibody. n = 3. H, ECs were transfected with scrambled, TRIM28, RBPJκ siRNA alone or TRIM28 combined with RBPJκ siRNA for 48 hours, then exposed to VEGF165 (20 ng/mL), and the scratch migration assay was performed. n = 3. *P < .05. I-K,ECs were transfected with scrambled or TRIM28 siRNA or TRIM28 siRNA combined with RBPJκ siRNA and then, subjected to angiogenic sprouting assay. The representative images were shown (I). Scale bar, 100 μm. The sprout number (J) and cumulative sprout length (K) per bead were quantified. Data from three independent experiments were included and analyzed.**P < .01. L-R, ECs were transfected with either scrambled or TRIM28 siRNA alone, or co-transfected with STAT3 siRNA for 48 hours. The mRNA expressions of VEGFR1 (L), VEGFR2 (M), Notch1 (N), UNC5B (O), CXCR4 (P), STAT3 (Q), and TRIM28 (R) were analyzed using qPCR method. n = 3. **P < .01.Statistical significance was calculated by Tukey’s multiple comparison test