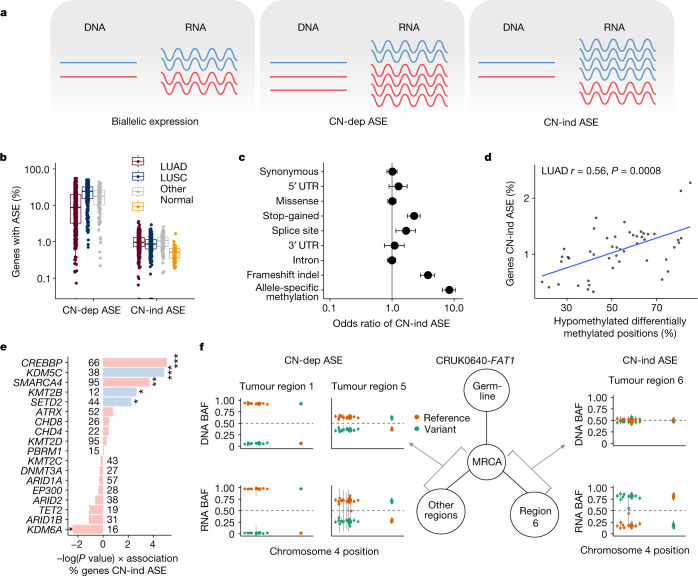
Fig. 2. ASE in NSCLC.
a, Schematic displaying the concepts of biallelic expression, CN-dependent ASE (CN-dep ASE) and CN-independent ASE (CN-ind ASE). b, Proportion of evaluable (containing an expressed SNP) genes affected by CN-dependent ASE and CN-independent ASE in tumours and normal tissue samples. LUAD, n = 454 regions from 144 tumours; LUSC, n = 293 regions from 88 tumours; Other, other subtypes, n = 130 regions from 38 tumours; Normal, tumour-adjacent normal lung tissue, n = 95. c, Points indicate odds ratio estimates for CN-independent ASE when somatic point mutations, or ASM (in samples for which both RRBS and RNA-seq were available) was concomitantly detected in the same gene, by type of alteration. Bars indicate 95% confidence intervals. Odds ratios for the links between CN-independent ASE and mutations and between CN-independent ASE and ASM are based on 876 primary tumour regions from 332 tumours and on 96 tumour regions from 31 tumours, respectively. d, Relationship in LUAD between the proportion of evaluable genes with CN-independent ASE and the ratio of differentially hypomethylated clusters of neighbouring CpGs compared to all differentially methylated genomic positions. The P value was calculated using a linear mixed-effects model with tumour as the random variable. e, Linear mixed-effects model showing the impact of driver mutations in candidate epigenetic modifier genes22 (mutated in more than five tumours) and tumour mutational burden on the proportion of evaluable genes with CN-independent ASE. Factors independently associated with increased CN-independent ASE in a multivariable model are coloured blue. *P < 0.05, **P < 0.01, ***P < 0.001. f, An example of genomic–transcriptomic mirrored subclonal allelic imbalance occurring in FAT1 within CRUK0640. DNA and RNA B allele frequencies (BAFs) for each SNP in FAT1 are plotted and coloured according to the reference and variant status of each allele for each region sampled within the tumour. In this instance, there is evidence of CN-dependent ASE in two regions and CN-independent ASE in one region. These events favour overexpression of different parental alleles and occur on different branches of the phylogenetic tree; a simplified version is displayed. MRCA, most recent common ancestor.