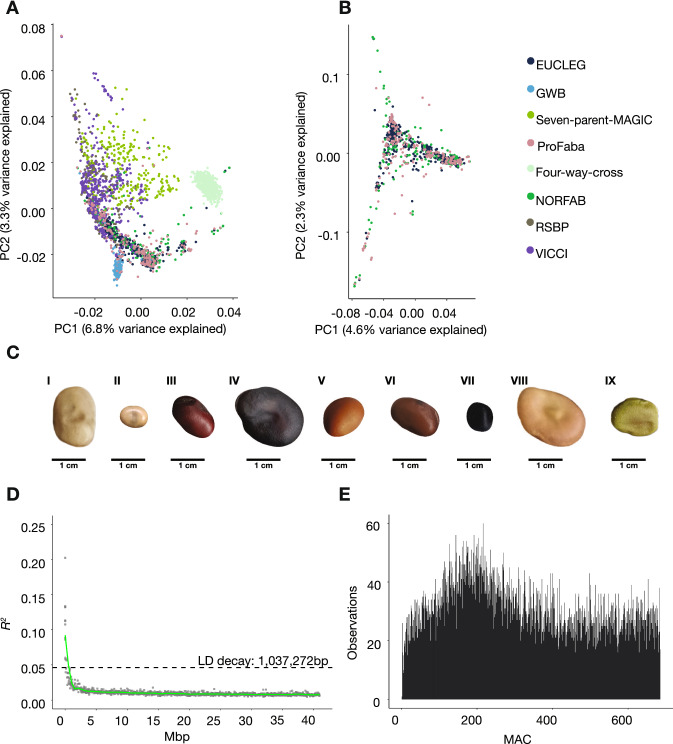
Fig. 3.
Characterization of the diversity panel. A, B The genetic structure of the data, as indicated by the first and second principal components and color-coded by panel membership A All panels, n = 2678. B The inbred EUCLEG, ProFaba, and NORFAB diversity panels, n = 787. C An image-based representation of the large phenotypic variation of seeds in the diversity panel. I) GPID_00080, II) EUC_VF_131, III) GPID_00162, IV) GPID_00176, V) GPID_00163, VI) GPID_00119, VII) GPID_00004), VIII) EUC_VF_272, IX) GPID_00042. D LD decay plot for the diversity panel. Y-axis displays the average squared correlation coefficient (R2) between markers when sorted after the average distance and binned into groups of 1000. For each bin, the x-axis displays the average distance in Mbp between two SNPs. The green line is the fitted loess curve with half its maximum R2 indicated by the dotted line. E Folded site-frequency spectrum of non-monomorphic SNPs in the diversity panel. The x-axis reports the minor allele counts (color figure online)