Fig. 4.
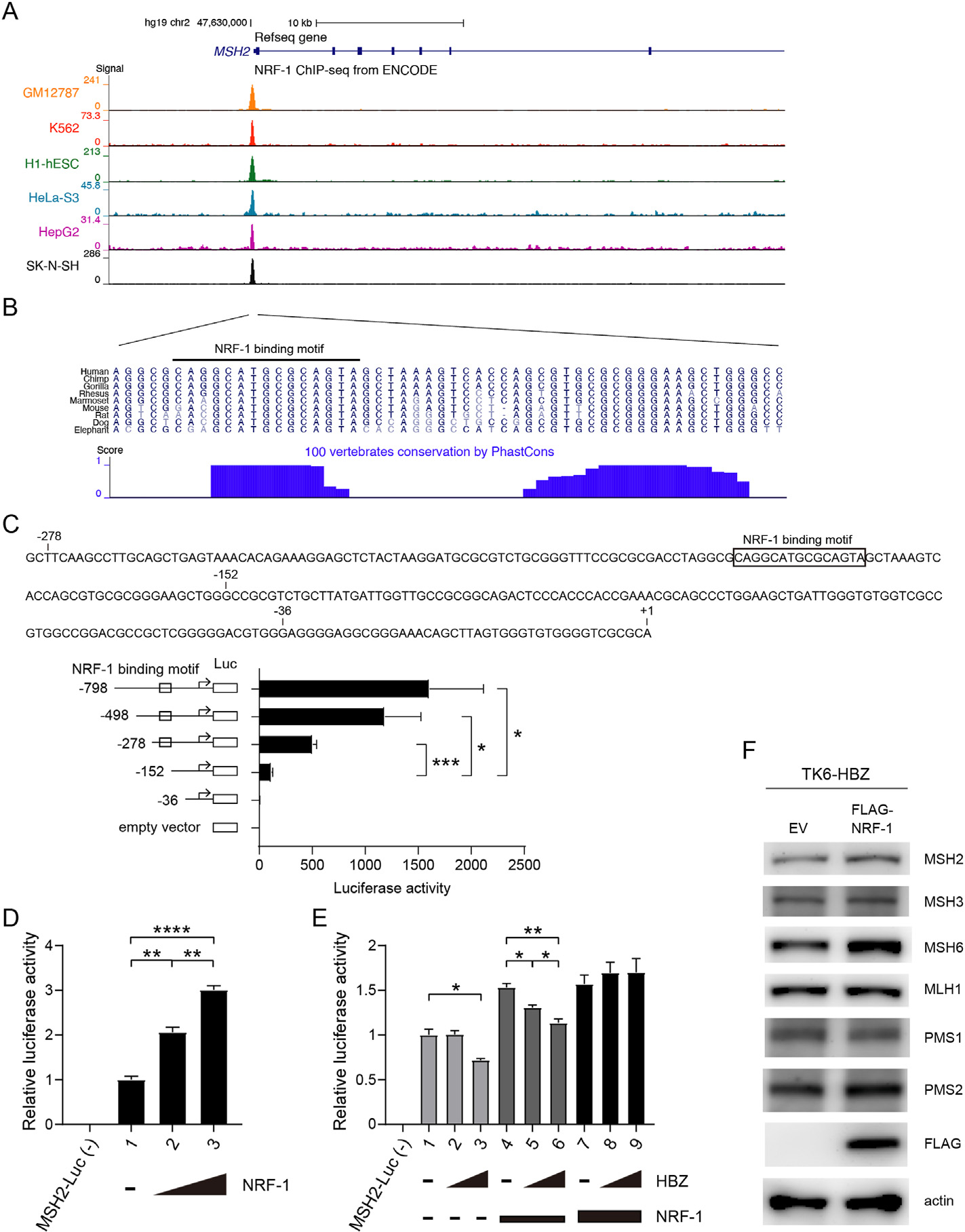
HBZ suppresses transcription of MSH2 via the transcription factor NRF-1 (A, B) UCSC genome browser view of the NRF-1 binding site in the MSH2 promoter. (A) Tracks show signals of NRF-1 ChIP-seq in the promoter region of MSH2 from the ENCODE project. (B) Sequences of the signal peak are shown. Blue bars represent the degree of sequence conservation across different vertebrates from the phastCons program. The black-lined region represents the NRF-1 binding motif predicted using Factorbook. (C) Activity of MSH2 promoter constructs truncated at different positions upstream of the transcription start site (TSS). Sequence upstream of the TSS of MSH2, and the NRF-1 binding motif predicted using Factorbook are shown. Each MSH2-Luc was transfected into HEK293T cells together with Renilla-Luc. MSH2-luciferase activity was assayed as relative to Renilla-Luc (n = 3, mean ± SEM). (D) Effects of NRF-1 expression on MSH2 promoter activity. MSH2-Luc and Renilla-Luc were transfected into HEK293T cells, with or without vectors expressing NRF-1 (0.05 μg or 0.2 μg) (n = 3, mean ± SEM). (E) Effects of HBZ on MSH2 promoter activity. MSH2-Luc and Renilla-Luc were transfected into HEK293T cells, with or without vectors expressing HBZ (0.4 μg or 0.6 μg) and NRF-1 (0.0125 or 0.025 μg) (n = 3, mean ± SEM). (F) Immunoblotting of the indicated proteins in TK6-HBZ cells infected with lentivirus expressing FLAG-tagged NRF-1 or the corresponding empty vector (EV).
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (unpaired Student’s t-test).
