Fig. 2 |. NMR-guided evolution of myoglobin.
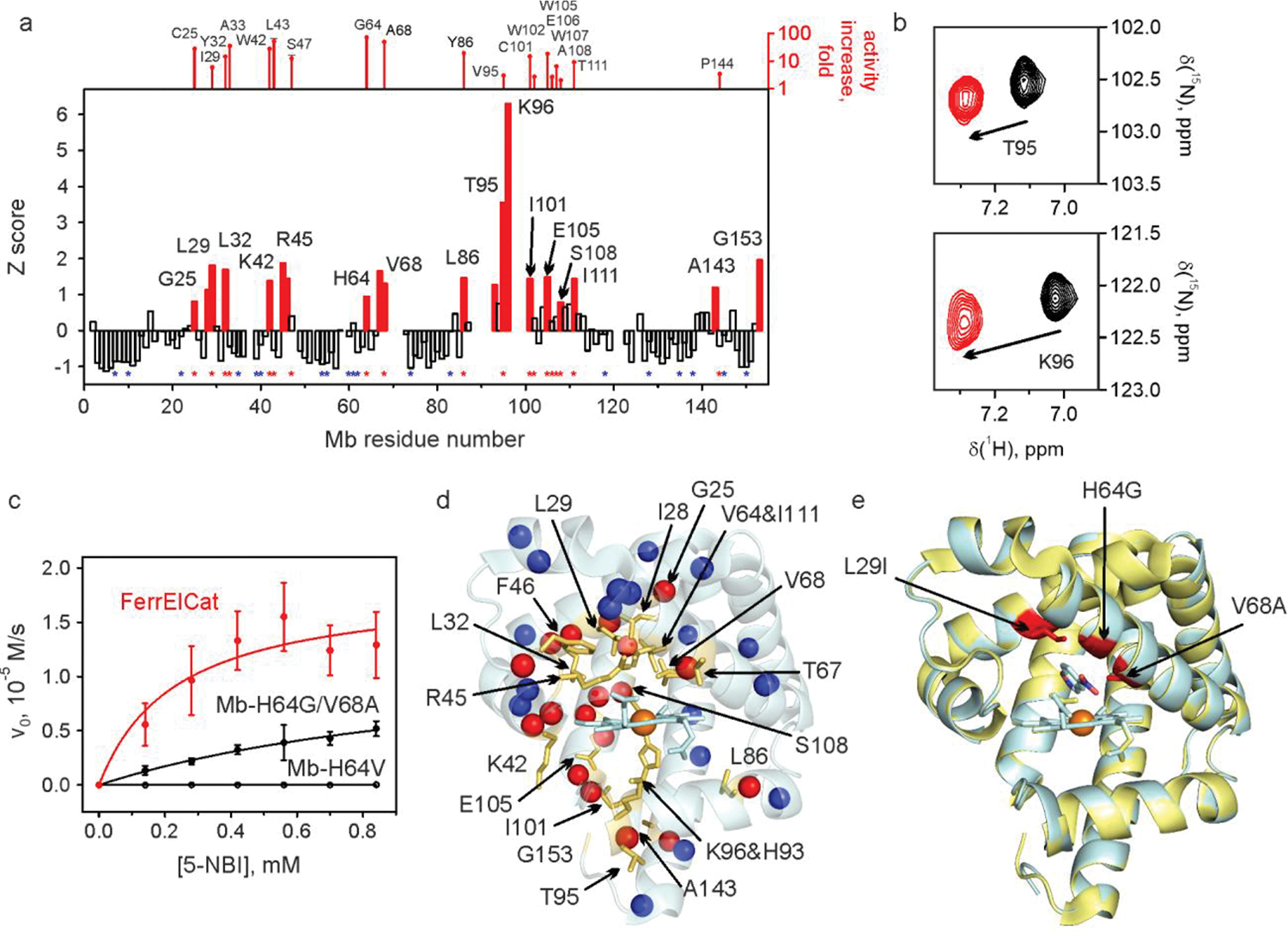
a, Backbone amide CSP of Mb-H64V upon addition of 2 molar equivalents of 6-NBT. The red bars indicate the protein regions experiencing large chemical shift perturbation (Z≳1). No bars are provided where no backbone resonance assignment could be made. The positions where productive mutations were found are marked by red asterisks (along with the corresponding increase in kcat/KM relative to Mb-H64V, top). Positions where screening identified no productive mutations are marked by blue asterisks. The corresponding representative 1H-15N HSQC spectral regions are shown in panel b. c, Michaelis-Menten plots for representative proteins. d, NMR CSP data mapped on the X-ray crystal structure of Mb-H64V (PDB 6cf0) showing the residues with prominent changes (Z≳1) as yellow sticks. The spheres show backbone nitrogen atoms of the residues with identified productive mutations (red) or those for which no productive mutations could be found (blue). e, Overlay of the crystal structures of Mb-H64V (yellow) and FerrElCat with the docked inhibitor (cyan). The newly introduced mutations are shown in red.
