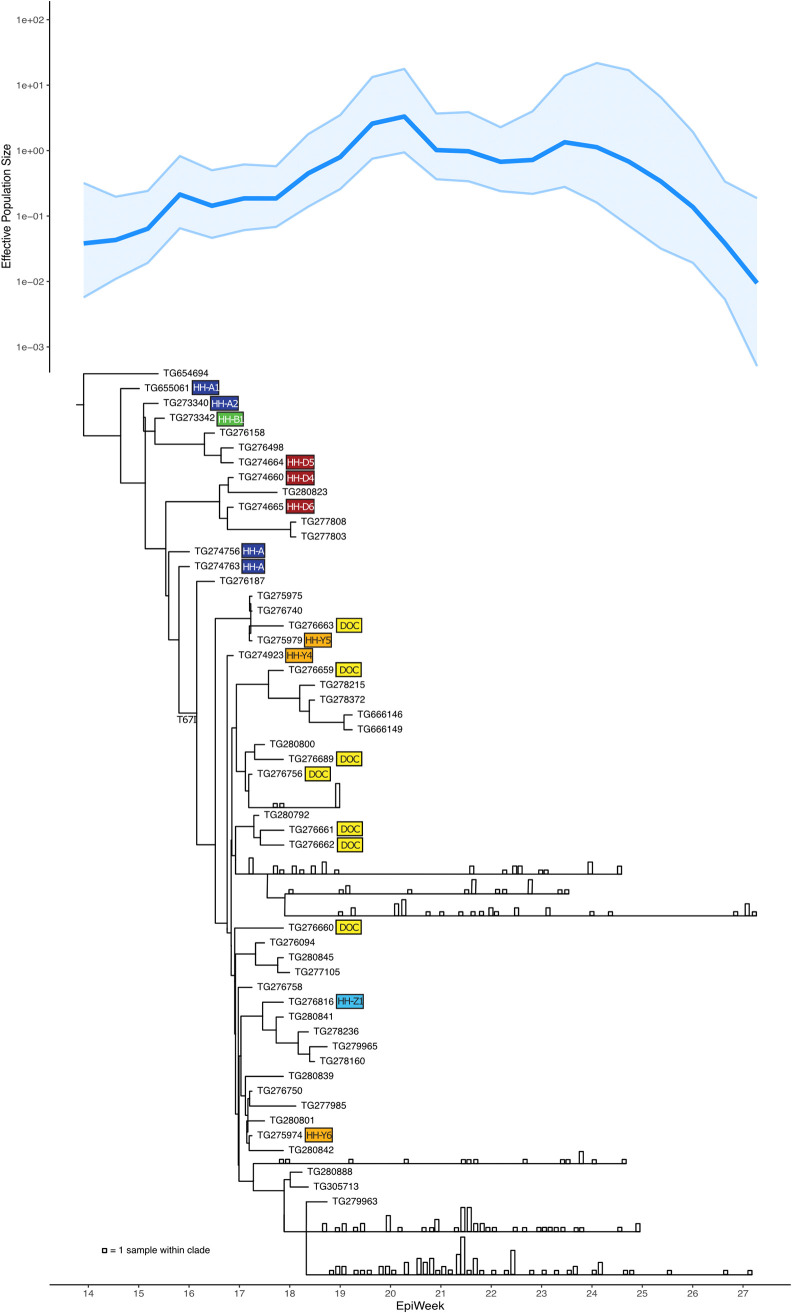
FIG 1.
Effective population size (Ne) analysis plot and phylogenetic tree of viral lineage B.1.289, characterized by spike: H245Y and Orf1b: T67I, representing the initial SARS-CoV-2 outbreak on an Arizona tribal reservation. This analysis includes 276 virus genomes, including the first known tribal case virus. Some large clades are collapsed, with vertical bars representing the number of samples in the clade collected on that date (see Fig. S1 for the full phylogenetic tree). The largely linear transmission pathway illustrated by the tree and the robust sampling and sequencing illustrated by the Ne plot reflect the unique circumstances of this lineage’s introduction and aggressive response by tribal public health. B.1.289 first appeared in April 2020, and its elimination was successful by July. Colored boxes denote SARS-CoV-2 cases associated with cluster 1 and 2, matching those depicted in the epidemiological linkage maps in Fig. 3.