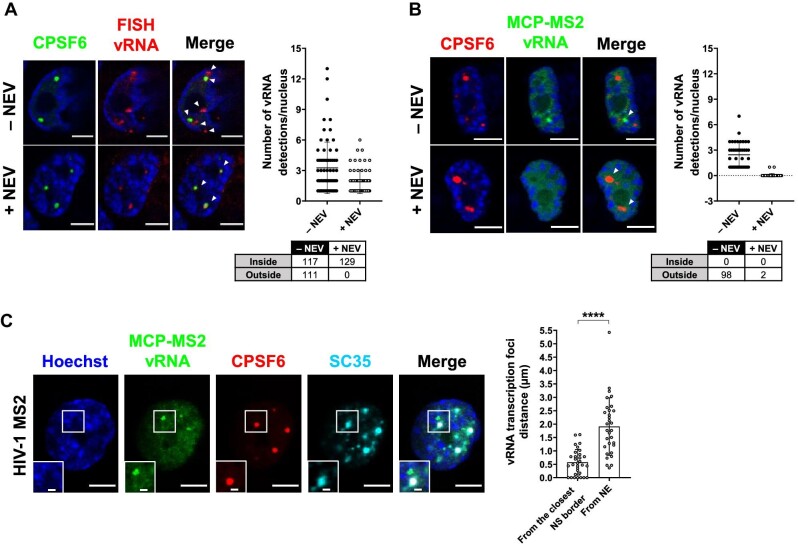
Figure 5.
MCP-MS2 RNA labelling system detects transcription foci only. (A) Confocal images of immuno-RNA FISH in THP-1 cells infected with HIV-1 and treated with or without 10 µM NEV (MOI = 20, 3 days p.i.). The graph shows the number of vRNA per nucleus, and the table shows the amount of vRNA inside or outside CPSF6 clusters (number of cells: 70 (–NEV), 68 (+NEV)). Two independent experiments were performed. (B) Confocal images of THP-1 cells expressing MCP-GFP infected with HIV-1 ANCH3 MS2 and treated with or without 10 µM NEV (MOI = 2.5, 3 days p.i.). The graph shows the number of vRNA per nucleus, and the table shows the amount of vRNA inside or outside CPSF6 clusters (number of cells: 40 (–NEV), 44 (+NEV)). Three independent experiments were performed. (C) Confocal images of THP-1 cells expressing MCP-GFP infected with HIV-1 ANCH3 MS2 (MOI = 2.5, 3 days p.i.). The graph shows the distance of each RNA focus from the closest NE point or SC35-marked NS border ± SD (31 vRNAs, 8 cells). Results of two independent experiments were analyzed from 3D acquisitions. Unpaired t-test, ****P ≤ 0.0001. Scale bar, 5 µm. Inset, 1 µm.