Figure 2.
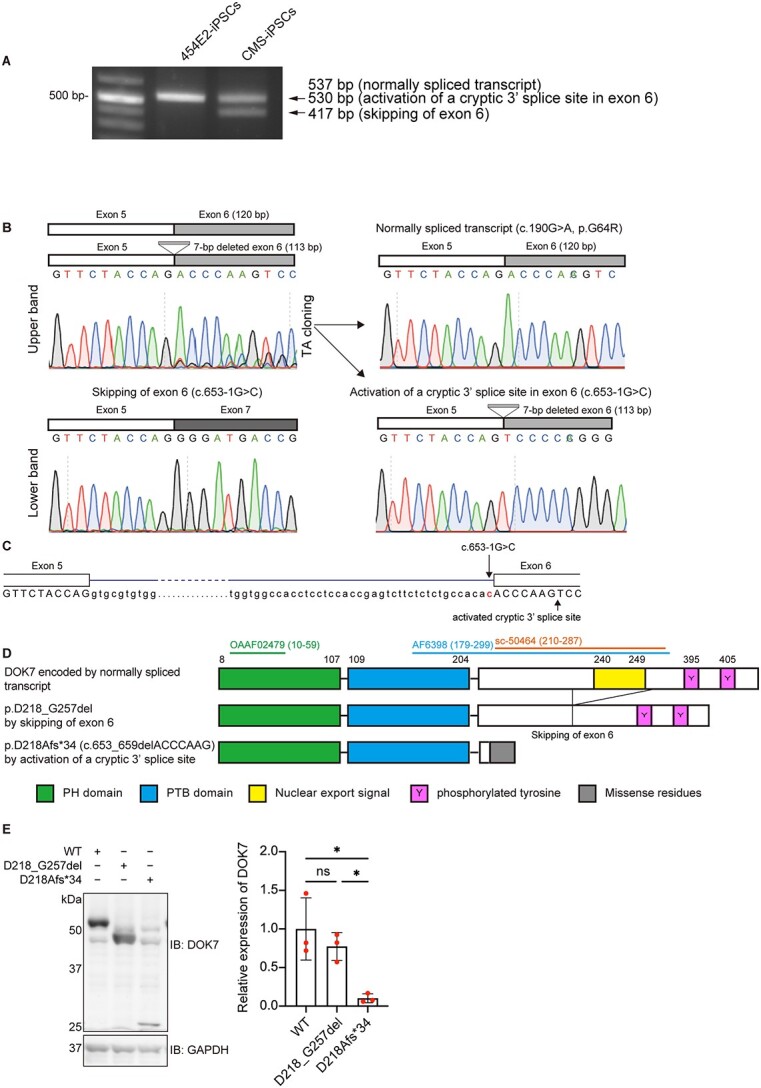
Aberrantly spliced transcripts due to c.653-1G > C. (A) RT-PCR spanning DOK7 exon 6 of control 454E2-iPSCs and patient-derived CMS-iPSCs. The upper band was comprised of 537-bp and 530-bp fragments. (B) Sequence chromatograms of three RT-PCR products in CMS-iPSCs. Chromatograms of the two transcripts in the upper band in A are indicated on the right. (C) Schematic showing the positions of the c.653–1 > C mutation and activated cryptic 3′ splice site that is seven nucleotides downstream of the intron 5/exon 6 junction. (D) Schematic presentation of DOK7 proteins encoded by three transcripts in B. Epitopes of three DOK7 antibodies (OAAF02479, AF6398 and sc-50464) used in this study are indicated with codon numbers in parentheses. See Supplementary Material, Table S2 for the details of these antibodies and Supplementary Material, Fig. S5A for representative western blotting. (E) Representative western blotting and quantitative analysis of mutant DOK7 arising from an allele with c.653-1G > C in transfected COS7 cells. Expression levels were normalized to that of GAPDH and to the ratio of wild-type (WT) DOK7. Mean and SD (n = 3 experiments) are indicated with individual values in red dots. One-way ANOVA with Dunnett’s post hoc multiple comparison test was applied (ns, no significance; *P < 0.05).
