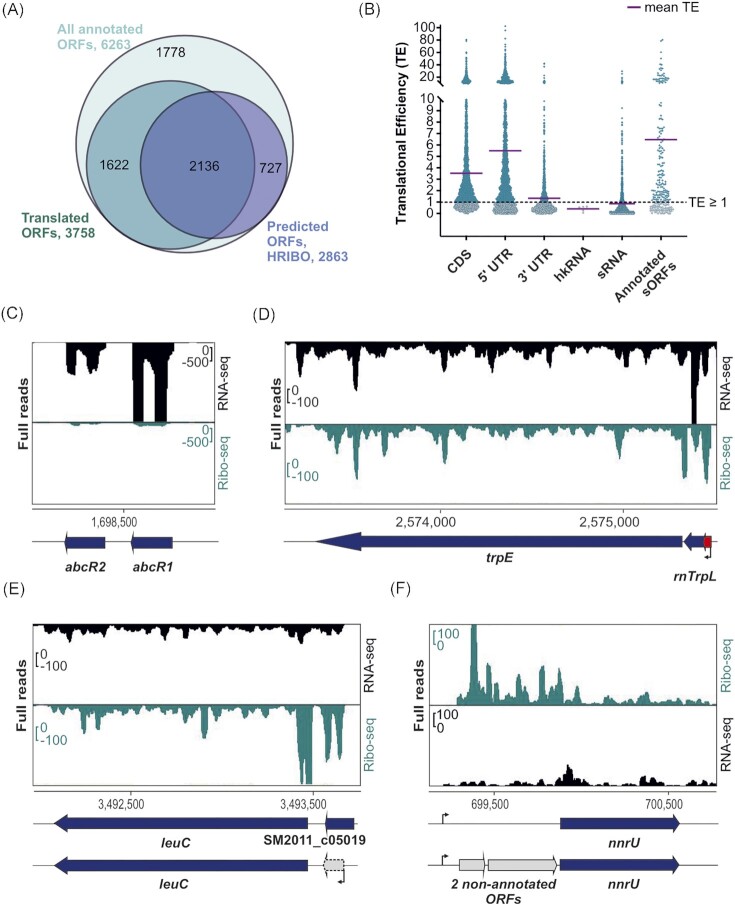
Figure 2.
Ribosome profiling (Ribo-seq) captures the translatome of Sinorhizobium meliloti 2011 and reveals some features at the single-gene level. (A) Comparison of all annotated open reading frames (ORFs), annotated translated ORFs detected by Ribo-seq, and ORFs predicted to be translated by tools included in the HRIBO pipeline. To detect translation, we used the following parameters on the Ribo-seq data: TE of ≥ 0.5 and RNA-seq and Ribo-seq RPKM of ≥ 10. The numbers of ORFs per category are shown and represented by area size. Diagrams were prepared with BioVenn (www.biovenn.nl). (B) Scatter plot showing global TEs (TE = Ribo-seq/RNA-seq) computed from S. meliloti Ribo-seq replicates for all annotated coding sequences (CDS), annotated 5'- and 3'-UTRs, annotated housekeeping RNAs (hkRNA), annotated small RNAs (sRNAs) with (putative) regulatory functions, and annotated sORFs encoding proteins of ≤ 70 amino acids (aa). The purple lines indicate the mean TE for each transcript class. (C) Analysis of the two well-characterized sRNAs AbcR1 and AbcR2 by Ribo-seq. These two sRNAs show read coverage mostly in the RNA-seq library. (D) Ribo-seq reveals the active translation of the trpE leader peptide peTrpL (14 aa, encoded by the leaderless sORF trpL in the 5'-UTR (red arrow) and/or by the attenuator sRNA rnTrpL). In addition, the coverage of the Ribo-seq library shows that the biosynthetic gene trpE is translated in minimal medium, as expected. (E) Re-annotation of sORF SM2011_c05019 (50 aa). The GenBank 2014 annotation does not fit the RNA-seq and Ribo-seq read coverages. HRIBO predicts a shorter leaderless sORF (38 aa) that corresponds to the read coverage in both libraries. (F) Two ORFs missing from the GenBank 2014 annotation are revealed by Ribo-seq upstream of the nnrU gene related to denitrification. Genomic locations and coding regions are indicated below the image. Bent arrows indicate transcription start sites based on (Sallet et al. 2013).