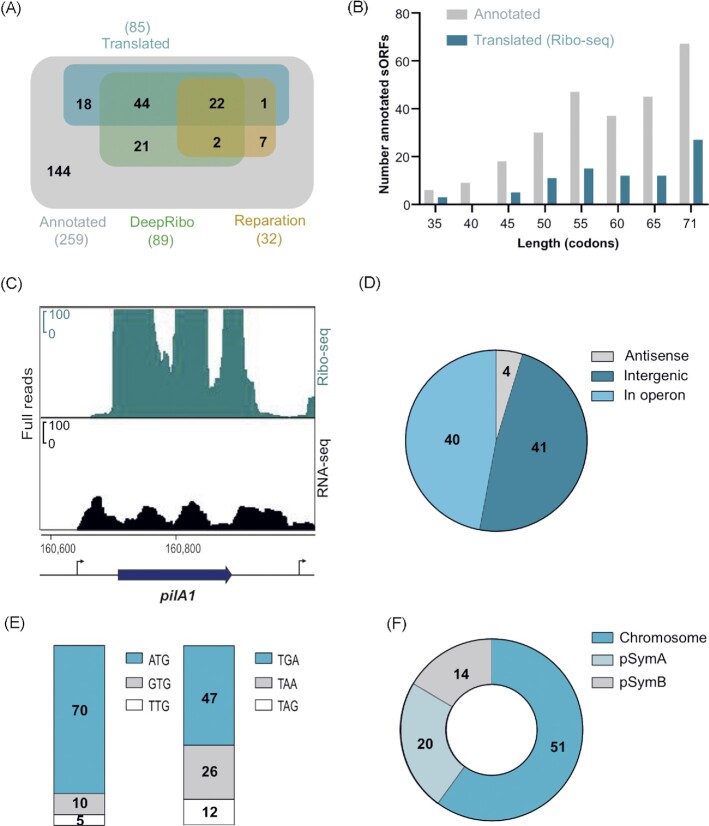
Figure 3.
Ribo-seq reveals translated annotated small open reading frames (sORFs) in Sinorhizobium meliloti 2011. (A) Venn diagrams showing the overlap between all annotated sORFs (259 sORFs, GenBank 2014), the sORFs detected as translated by Ribo-seq (benchmark set, TE of ≥ 0.5, RNA-seq and Ribo-seq RPKM of ≥ 10, and extensive manual curation), and sORFs predicted by the automated ORF prediction tools Reparation or DeepRibo. (B) Histogram showing the length distribution of the 85 annotated sORFs identified as translated by Ribo-seq in comparison with the 259 annotated sORFs. (C)Integrated genome browser screenshot depicting reads from the Ribo-seq and RNA-seq libraries for the annotated sORF pilA1 (60 amino acids, encoding a pilin subunit). The genomic position and the coding region are indicated below the image. Bent arrows indicate transcription start sites based on (Sallet et al. 2013). (D) Genomic context for the translated annotated sORFs relative to the annotated neighboring genes. (E) Start (left) and stop (right) codon usage of the translated annotated sORFs. (F) Replicon distribution of the translated annotated sORFs.