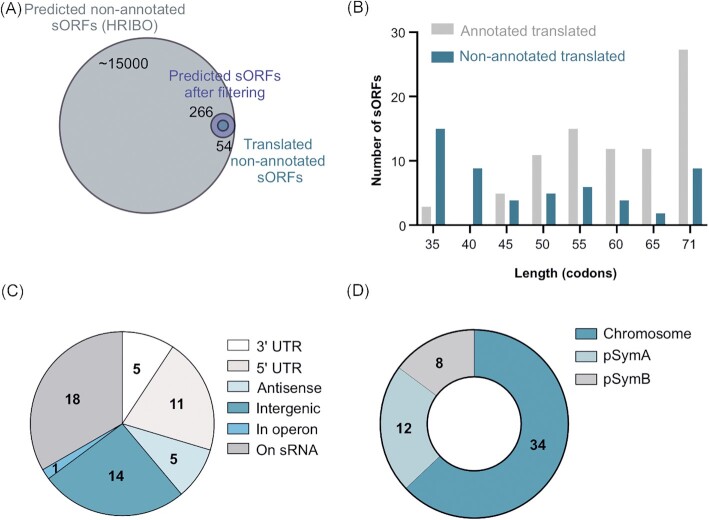
Figure 4.
Ribo-seq uncovers a repertoire of small open reading frames (sORFs) missing from the Sinorhizobium meliloti 2011 genome annotation. (A) sORF predictions from HRIBO included a high number of potential non-annotated sORFs (approximately 15,000). These sORFs were first filtered (TE of ≥ 0.5, RNA-seq and Ribo-seq RPKM of ≥ 10, DeepRibo score of > −0.5) to generate a set of 266 translated sORF candidates that were additionally manually curated by inspection of the Ribo-seq read coverage in a genome browser. Overall, 54 high-confidence non-annotated sORFs displayed translation during growth in minimal medium. A Venn diagram shows the respective number of proteins from each category (scaled with area size). Diagrams were prepared with BioVenn ( www.biovenn.nl). (B) Histogram showing the length distribution of the 54 non-annotated versus the 85 annotated sORFs identified as translated by Ribo-seq. (C) Genomic context of the translated non-annotated sORFs. (D)Replicon distribution of the translated non-annotated sORFs.