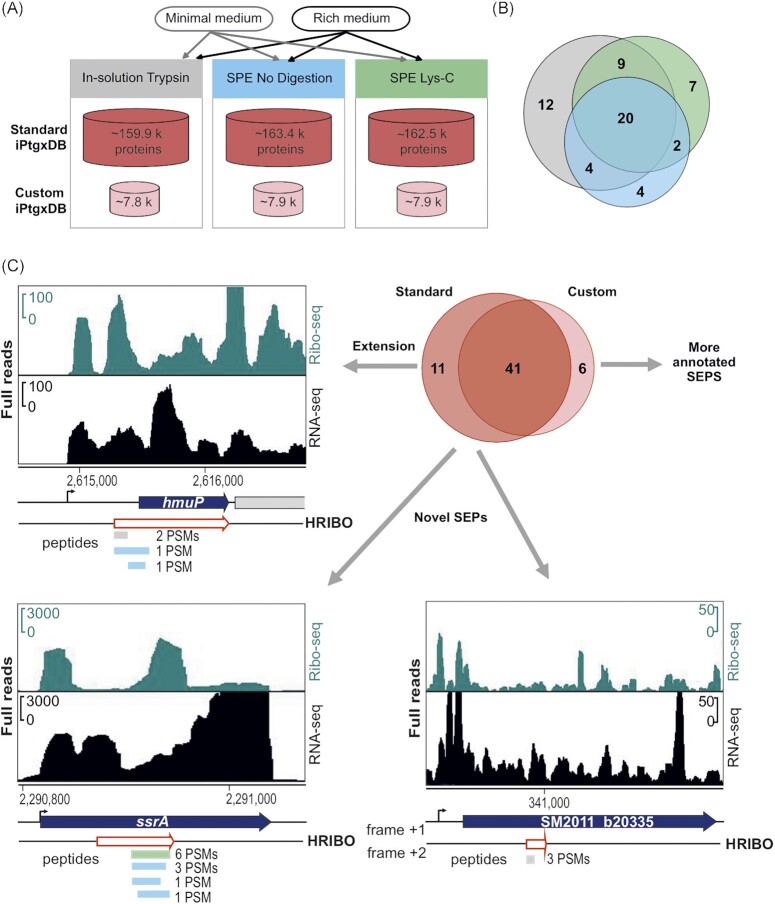
Figure 5.
Mass spectrometry-based identification of known and novel small open reading frame-encoded proteins (SEPs). (A) Experimental set-up for the proteomics analyses. Bacteria were grown in minimal and rich media, and protein extracts were further processed with tryptic in-solution digest (gray), solid-phase enrichment (SPE) of small proteins with subsequent Lys-C digestion (green), or without further digestion (blue). (B) Overlap of the identified SEPs by experimental approach; trypsin identified 45 SEPs; compared with the trypsin approach, Lys-C identified 38 SEPs (nine novel, 24%), and the approach without digestion found 30 SEPs (six novel, 20%). (C) Novel/unique identifications uncovered by the standard integrated proteogenomic search databases (iPtgxDB) and the small custom iPtgxDB. Standard iPtgxDB: Three peptides imply a 14 aa longer proteoform (60 aa) for HmuP than annotated; four peptides of the tmRNA-encoded proteolysis tag were identified; one peptide (3 peptide spectrum matches [PSMs]) implied a novel SEP (34 aa) internal to the genomic region that also encodes SM2011_b20335 but in a different frame. Spectra identifying these peptides are shown in Fig. S5. These identifications were also predicted by HRIBO based on Ribo-seq. Finally, six annotated proteins (GenBank 2014 and/or RefSeq 2017) were identified only in the search against the small custom iPtgxDB, as they did not accumulate enough spectral evidence in the search against the standard iPtgxDB (Table S4).