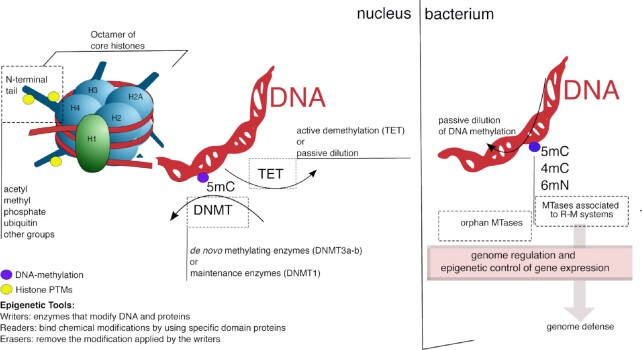
Figure 1.
The epigenomes of eukaryotes and bacteria. In eukaryotes, epigenetic modifications involve DNA methylation (purple tag) and histone modifications [yellow tag; for a complete list of possible chemical modifications on histone proteins please refer to Huang et al. (2014) and Zhao and Garcia (2015)]. The DNA is packaged in a structure called chromatin, which regulates its activity and inheritance and is organized in fundamental structures called nucleosomes. Each nucleosome consists of a segment of 147 bp of DNA wrapped around an octamer of proteins containing two copies each of four different histones: H2A, H2B, H3, and H4. This arrangement of 11 nm of DNA and its associated proteins forms a fiber, which plays a major role in the cell. In fact, the regulatory proteins that interact with target subunits of the fiber can increase or decrease the compactness of the chromatin structure, leading to enhancing or reducing gene expression. Separate enzymes are responsible for de novo methylation and the maintenance of DNA methylation. DNA demethylation can occur by an active process driven by dedicated proteins (TET enzymes), or by a passive one, where DNA methylation is diluted upon DNA replication. Typically, the methylated base in eukaryotes is C5m, whereas it is often N6m in bacteria. Most bacterial DNA-methyltransferases (MTases) belong to the restriction–modification (R–M) system, responsible for genome defense, whereas orphan DNA MTases have no apparent cognate REase. Both kinds of MTases play a role in epigenetic regulations in bacteria.