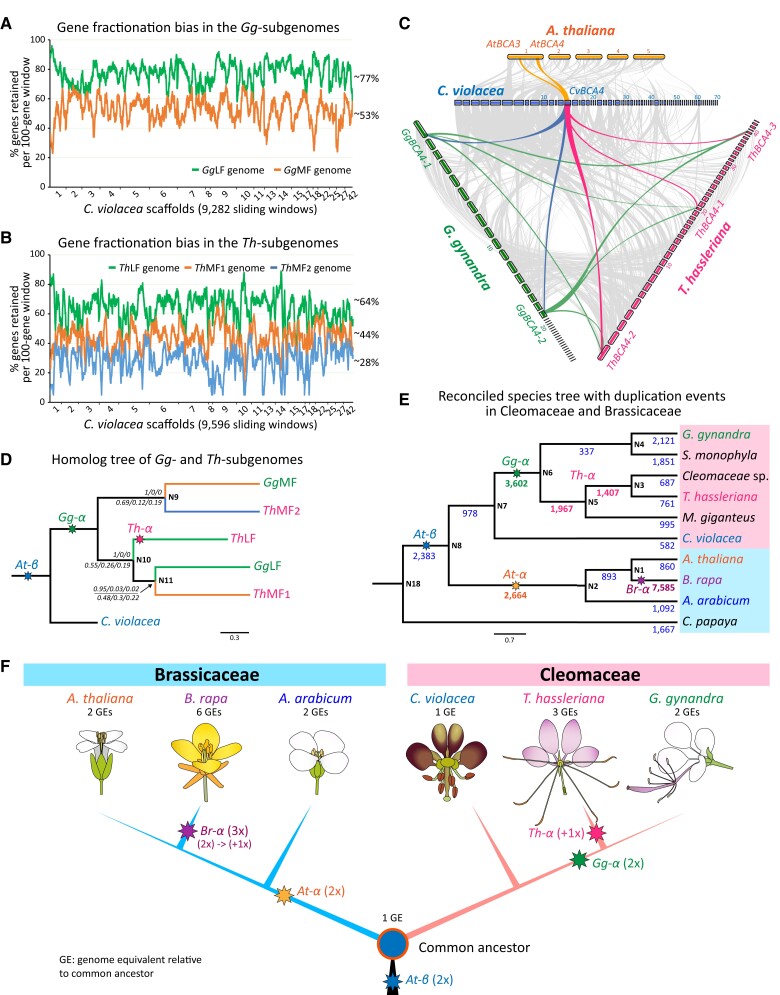
Figure 3.
Subgenome fractionation bias and phylogenetic relationship of G. gynandra, Cleomaceae and Brassicaceae species. Gene fractionation bias in the two Gg-subgenomes, GgLF and GgMF of G. gynandra (A) and three Th-subgenomes, ThLF, ThMF1 and ThMF2 of T. hassleriana (B). For both genomes, the C. violacea genome was used as reference. Most of the synteny was detected within the largest 20 Cv scaffolds. Gene retention (%) was calculated for each sliding window of 100 genes across the Cv scaffolds. C, Macrosynteny and microsynteny patterns show that a genome region bearing gene BCA4 (BETA CARBONIC ANHYDRASE4) in the C. violacea genome can be tracked to two regions in A. thaliana (yellow lines), two regions in the G. gynandra genome (blue lines), and three regions in the T. hassleriana genome (red lines). The background grey wedges highlight major syntenic blocks (minspan = 30 genes) between genomes. D, Phylogenetic relationships of subgenomes of G. gynandra and T. hassleriana. The tree was rooted using the C. violacea genome as outgroup. Supporting values at each node are posterior probability (upper) and quartet scores (lower). Tree was constructed using the species-tree approach based on 52 genes located on four syntenic blocks that were found across three species and their subgenomes. Branch length is in coalescence units. E, A reconciled species-tree of Cleomaceae and Brassicaceae species with their duplication events. The numbers provided at each node correspond to the gene duplications detected for each clade. Genome data from A. arabicum, A. thaliana, B. rapa, C. violacea, G. gynandra, T. hassleriana, and C. papaya; and transcriptome data of other species were used. Tree topology was adapted from the ASTRAL-III coalescent-based species phylogeny (Mabry et al., 2020). All branch supporting values are >0.7 posterior probability, and not shown. Branch length is in coalescence units. Tree was rooted using C. papaya as outgroup (see Methods for more information). “N” in (D and E) denotes “tree node”. F, Phylogenetic relationships between Brassicaceae and Cleomaceae species/genera and ancient polyploidy events detected in both lineages: the At-β (blue star) shared by Brassicaceae and Cleomaceae; the At-α (yellow star) shared by all Brassicaceae, the Br-α event (purple star) in Brassica spp.; in Cleomaceae, the Gg-α (green star) shared by G. gynandra and T. hassleriana and a potential genome addition (red star) in T. hassleriana explaining the Th-α triplication observed in the species.