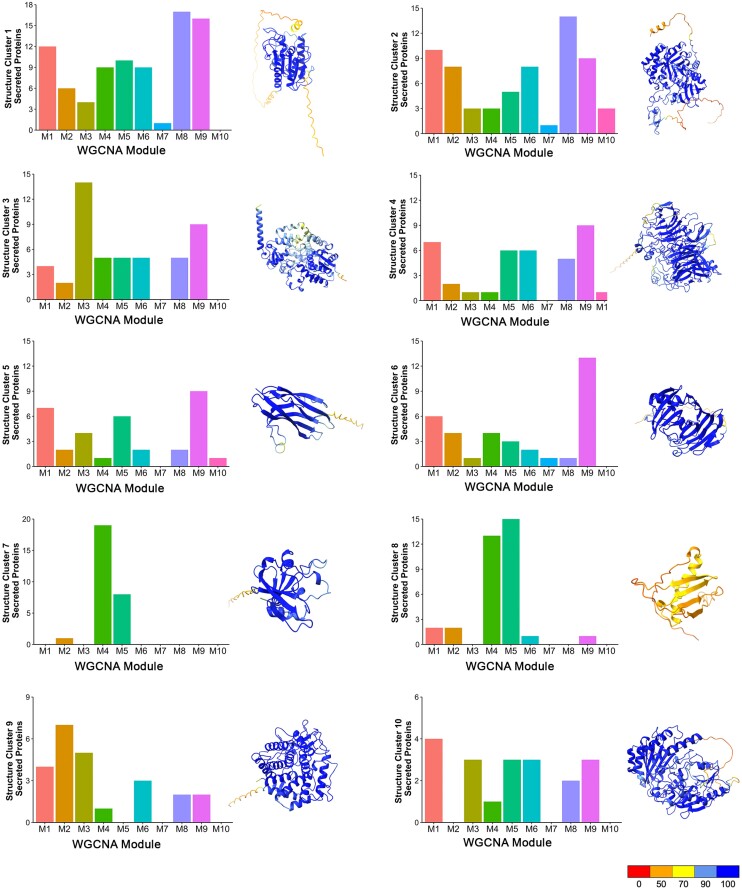
Figure 4.
Structurally conserved M. oryzae effectors are temporally co-expressed during biotrophic invasive growth. AlphaFold and ChimeraX platforms were used to predict the 3D structures of predicted secreted proteins of M. oryzae. Structure clusters were then analyzed against each WGCNA module. Bar charts represent the number of predicted secreted proteins identified in each predicted structure cluster (Clusters 1–10), which are classified within each WGCNA co-expression module during pathogenesis. Ribbon diagrams show a representative predicted protein structure for each M. oryzae structure cluster. These include MGG_09840, which contains a cutinase domain for Structure Cluster 1; MGG_04534 contains a chitin recognition protein domain for Structure Cluster 2; MGG_07497 contains a cytochrome P450 domain for Structure Cluster 3; MGG_05785 (Inv1/Bas113) contains a glycosyl hydrolase family 32 domain for Structure Cluster 4; MGG_02347 (Nis1) for Structure Cluster 5; MGG_08537 contains a glycosyl hydrolase family 12 domain for Structure Cluster 6; MGG_00230 (Mep2) is a predicted ART containing a heat-labile enterotoxin alpha chain domain for Structure Cluster 7; MGG_12426 is a predicted MAX effector and homologue of AvrPib for Structure Cluster 8; MGG_04305 contains a glycosyl hydrolase family 88 domain for Structure Cluster 9; MGG_00276 contains berberine and berberine-like domains for Structure Cluster 10. Protein structures were predicted by AlphaFold, and ChimeraX was used to visualize the protein structure using PDB files generated by AlphaFold. The command “color bfactor palette alphafold” was used to color-code the confidence for each prediction (scale from red = 0% confident to blue = 100% confident). Genes encoding proteins in Structure Clusters 7 and 8 are over-represented in WGCNA M4 and M5.