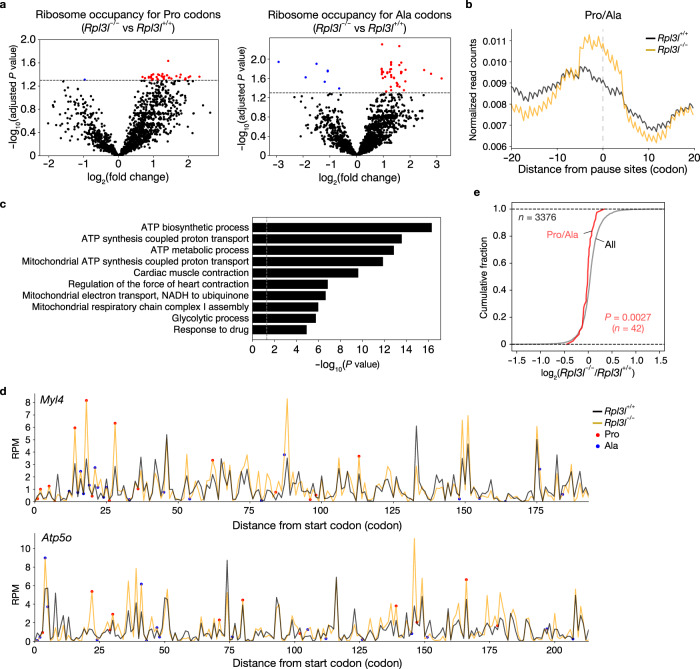
Fig. 5. Decreased protein expression associated with aberrant translation elongation dynamics in the RPL3L-ribosome–deficient heart.
a Volcano plots of differential ribosome occupancy for Pro and Ala codons in the heart of Rpl3l−/− mice compared with that of control mice (n = 4 mice). P values were calculated by two-sided Student’s t test and adjusted by Benjamini-Hochberg method for multiple comparisons, and the gray dashed lines indicate an adjusted P value of 0.05. b Metagene plots for the regions surrounding Pro/Ala sites with altered elongation dynamics for footprints of Ribo-seq analysis performed with the heart of Rpl3l+/+ or Rpl3l−/− mice (n = 4 mice). The average of replicates is shown. c GO analysis of genes with significantly altered elongation dynamics at Pro or Ala codons. P values were calculated as EASE scores, modified Fisher’s exact P values. The 10 GO terms with the smallest P values are listed. d Distribution of ribosome footprint occupancy at the A-site along the coding regions of Myl4 and Atp5o mRNAs in the heart of Rpl3l+/+ and Rpl3l−/− mice (n = 4 mice). The average of four replicates is shown. RPM, reads per million reads. e Cumulative fraction for fold change in protein expression in the heart of Rpl3l−/− mice compared with that of control mice (n = 4 mice). The expression level of proteins was analyzed by DIA-based MS, and the results are shown for all proteins (n = 3376) and for those encoded by the genes with differential ribosome occupancy at Pro or Ala codons. The P value was calculated by the two-tailed Mann-Whitney U test. Source data are provided as a Source Data file.