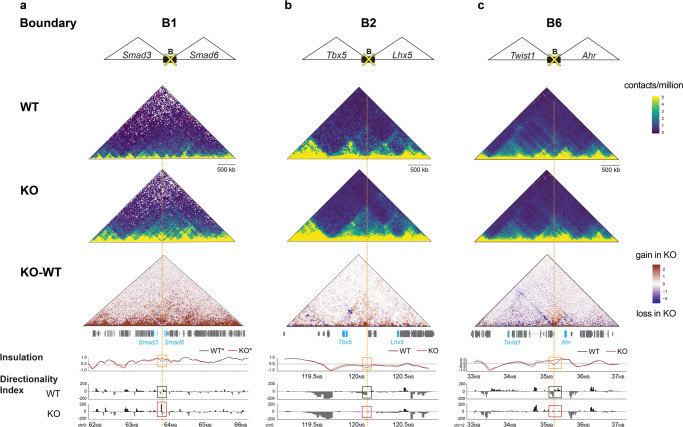
Fig. 3. Boundary deletions result in abnormal TAD architecture.
Hi-C derived interaction maps for TAD boundary loci B1 (a), B2 (b), and B6 (c). Cartoon of the TAD boundary deleted, with select developmental genes within the TADs flanking the deleted boundary (B), followed by three heatmaps showing Hi-C contact data. Heat maps (yellow-blue color-code) show Hi-C contact matrices presented as observed/expected contacts at 25 kb resolution in representative wild-type (WT) and knockout (KO) mouse liver tissue samples. For TAD boundary locus B1, note that the WT represents the wild-type allele in Cast background and KO represents the deletion allele in FVB background from an animal heterozygous for the TAD boundary deletion. The third heatmap (red-blue color-code) shows net changes in Hi-C interaction frequencies in the KO relative to WT. Positions of genes within the corresponding locus are indicated along the heatmap. The dashed orange vertical line indicates the position of the deleted boundary. Insulation profiles for the WT and KO samples corresponding to the heat maps are shown. The insulation profile assigns an insulation score to each genomic interval50, with local minima representing the most insulated region. Note the deviation from the minima in the insulation profile for the KO compared to WT (orange box), indicating loss of insulation in the KO. Bar plots show the Directionality Index (DI)5 for the same samples. Boundaries are called at regions where abrupt and significant shifts in upstream and downstream contacts are observed, as depicted in the WT (black box). Note either gain in contacts or the loss of demarcation between upstream and downstream contacts at the deleted site in the KO (red box). Genome coordinates shown are in mm10. More details on mouse strain background where applicable, replicates and additional TAD boundary deletions are provided in Supplementary Fig. 4 and “Methods”.