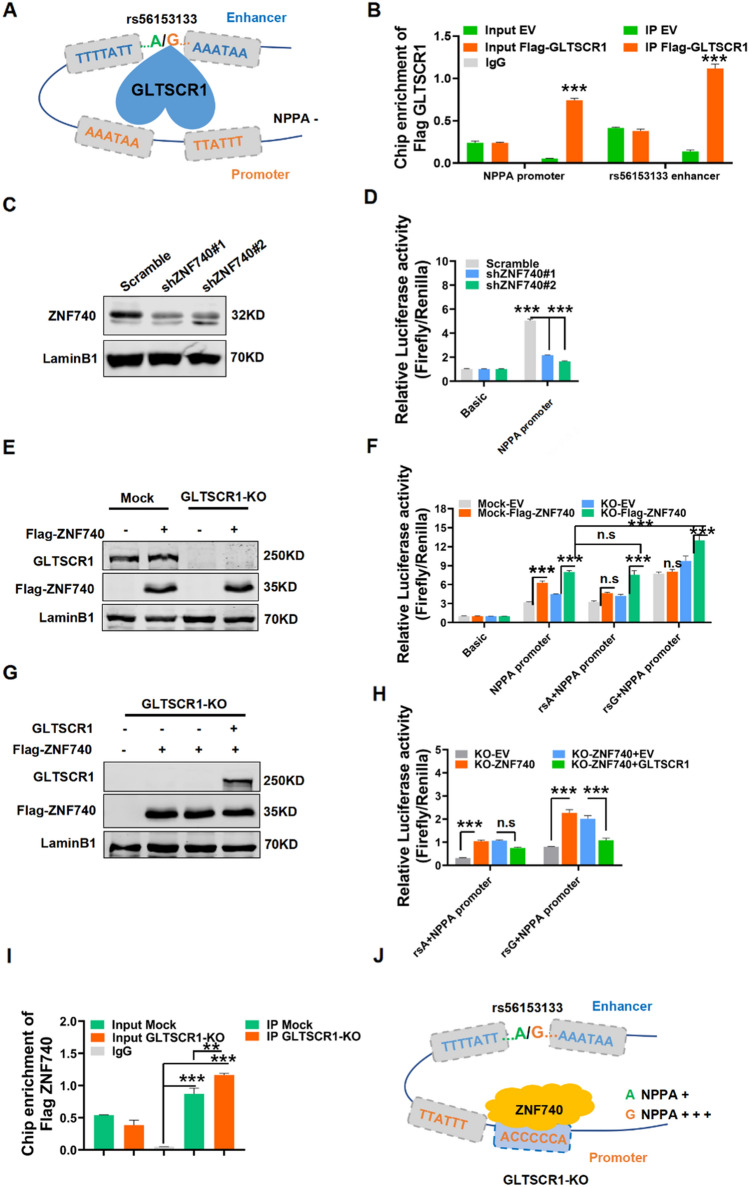
Fig. 7.
GLTSCR1 inhibits NPPA expression by blocking the interaction of ZNF740 in NPPA promoter. A The regulation model of GLTSCR1 to NPPA through binding NPPA enhancer and promoter. B Chip-qPCR analysis of GLTSCR1 binding at the NPPA promoter and the rs56153133 enhancer locus in HEK293T cells by anti-Flag beads. C Western blotting detected ZNF740 knockdown by shRNA in HEK293 cells. D Luciferase reporter assays using vectors containing the NPPA promoter to detect the transcriptional activity in ZNF740 knockdown cells. Luciferase signals were normalized to Renilla signals (n = 3). E Western blotting detected the overexpression of ZNF740 in HEK293 mock or GLTSCR1-KO cells. F Luciferase reporter assays using vectors containing the NPPA promoter or NPPA promoter with SNP locus enhancers to detect transcriptional activity in ZNF740-overexpressing mock or GLTSCR1-KO cells. Luciferase signals were normalized to Renilla signals (n = 3). G Western blotting detected ZNF740 overexpression and GLTSCR1 re-expression in GLTSCR1-KO HEK293 cells. H Luciferase reporter assays using vectors containing the NPPA promoter with the A or G allele of the rs56153133 SNP locus to detect the transcriptional activity of ZNF740 overexpression and GLTSCR1 re-expression in GLTSCR1-KO HEK293 cells. Luciferase signals were normalized to Renilla signals (n = 3). I Chip-qPCR analysis of ZNF740 binding at the NPPA promoter in mock and GLTSCR1-KO cells by anti-Flag beads. J The regulation model of ZNF740 to NPPA through binding NPPA promoter when GLTSCR1 is knockout. *P < 0.05, **P < 0.01, ***P < 0.001 compared to control (two-tailed Student’s t test were used)