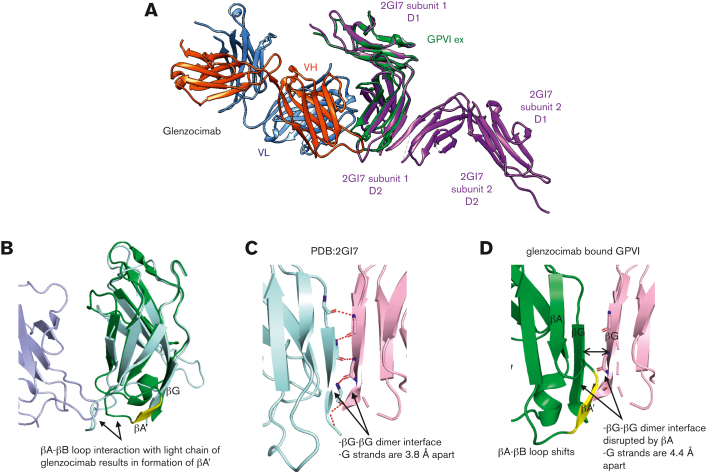
Figure 3.
Structural comparison between GPVIex/glenzocimab and 2GI7 crystals. (A) Overview of the superimposed dimeric GPVIex (PDB ID: 2GI7) on the GPVIex/glenzocimab complex (PDB ID: 7R58). The heavy chain (VH-CH1) of glenzocimab is depicted in red and the light chain (VL-CL) in blue. GPVI domain 2 (D2) bound to glenzocimab is represented in green and GPVI without any ligand (PDB ID: 2GI7) in purple. (B) Close up view of the overlayed D2 domains of dimeric GPVI (PDB:2GI7) (pale cyan) and glenzocimab-bound GPVI (green) highlighting the rearrangements within D2 because of clashes between the βA-βB loop of GPVI and the light chain of glenzocimab (light blue). Contacts with the light chain of glenzocimab cause a conformational change βA-βB loop resulting in the formation of a new β-strand, βA′, (shown in yellow). (C) Zoomed-in view of the dimeric interface formed between 2 βG strands from 2 GPVI subunits in the dimeric crystal structure (PDB:2GI7). Subunits are colored pale cyan and light pink, respectively. Polar contacts between each strand are shown as red dashed lines with binding resides shown as sticks. (D) Zoomed-in view of the βG strand of glenzocimab-bound GPVI (green) superimposed with the alternate GPVI subunit found in the dimeric structure (light pink). The conformational changes in the glenzocimab-bound D2 domain result in the formation of a new βA′ strand, which blocks the interaction between the 2 βG strands. The formation of βA′ also results in a shift in the βG strand resulting in an increased gap of 0.6 Å between the 2 βG strands. Combined, these factors explain how GPVI dimerization is blocked by glenzocimab.