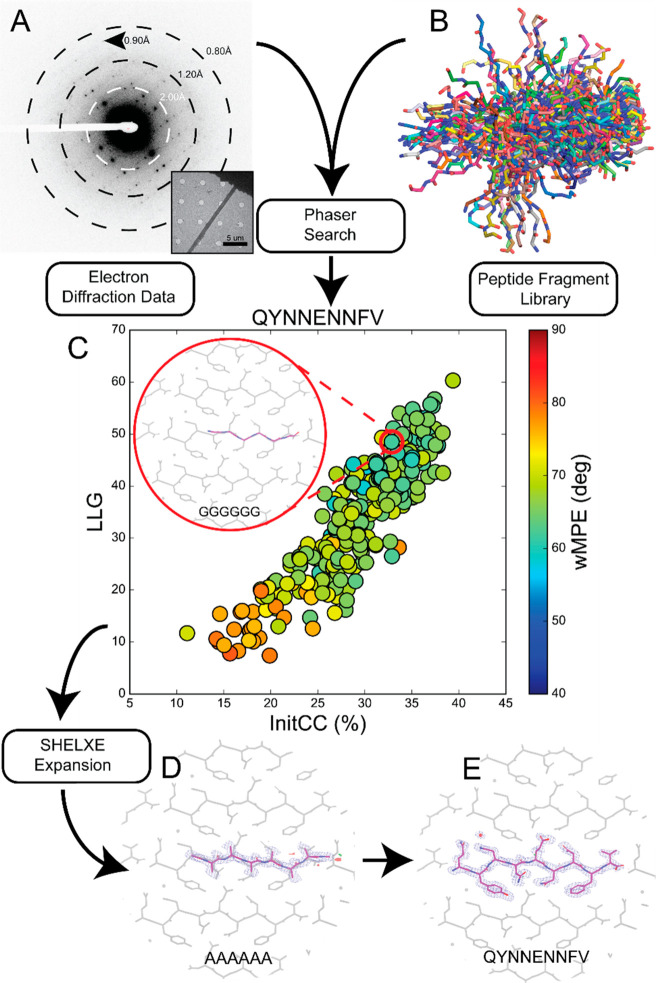
Figure 1.
Workflow for using peptide fragments for phasing MicroED data. A) Electron diffraction pattern reaching atomic resolution for QYNNENNFV. Rings designate resolution ranges while arrowhead designates highest resolution spot. B) All fragments comprising the polyglycine hexapeptide library aligned in pymol. C) LLG vs InitCC plot for the fragments screened in ARCIMBOLDO-BORGES. Color indicates the wMPE of the fragment relative to the phases calculated from the final structure. Inset shows the fragment chosen for SHELXE expansion overlaid on the final structure. D) Output solution from ARCIMBOLDO-BORGES following SHELXE expansion is shown overlaid on the final structure. Maps are shown after one round of refinement in Phenix. E) Final structure and potential map for the QYNNENNFV peptide with symmetry related chains shown in gray.