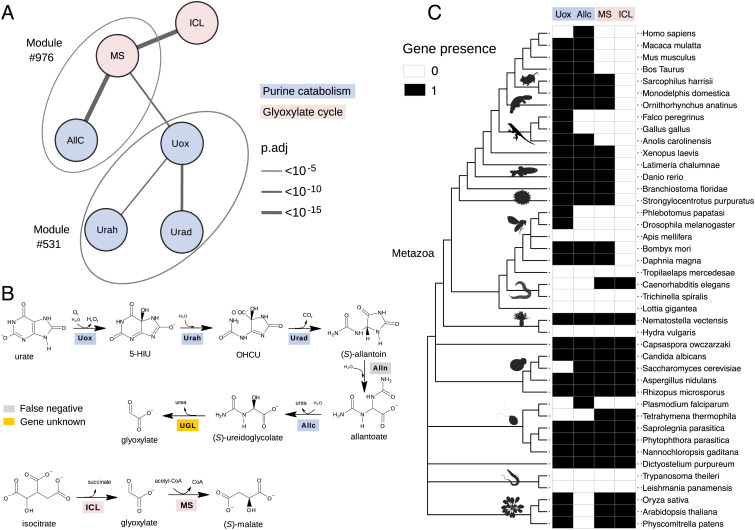
Fig. 4.
Identification of a missing gene in purine catabolism through cotr analysis. (A) Schematic network showing significant cotr associations of orthogroups representing urate oxidase (Uox) and allantoicase (Allc) with malate synthase (MS); other significant associations involve isocitrate lyase (ICL), HIU hydrolase (Urah), and OHCU decarboxylase (Urad). Modules defined by mcl clustering are encircled by ovals. (B) Scheme of the purine degradation and glyoxylate shunt reactions with the corresponding enzymes: True-positive genes retrieved by cotr analysis are shaded as in A; the false-negative allantoinase (Alln), member of the dihydropyrimidinase multigene family, is shaded gray; ureidoglycolate lyase (UGL) encoded by an unknown gene in Metazoa is shaded yellow. (C) Distribution map of Uox, Allc, MS, and ICL orthogroups across the ncbi phylogeny of selected eukaryotic species with emphasis on Metazoa. PhyloPic silhouettes (https://www.phylopic.org) are added to selected branches to aid species identification. A less detailed diagram of the gene distribution in 1,264 species is shown in SI Appendix, Fig. S13.