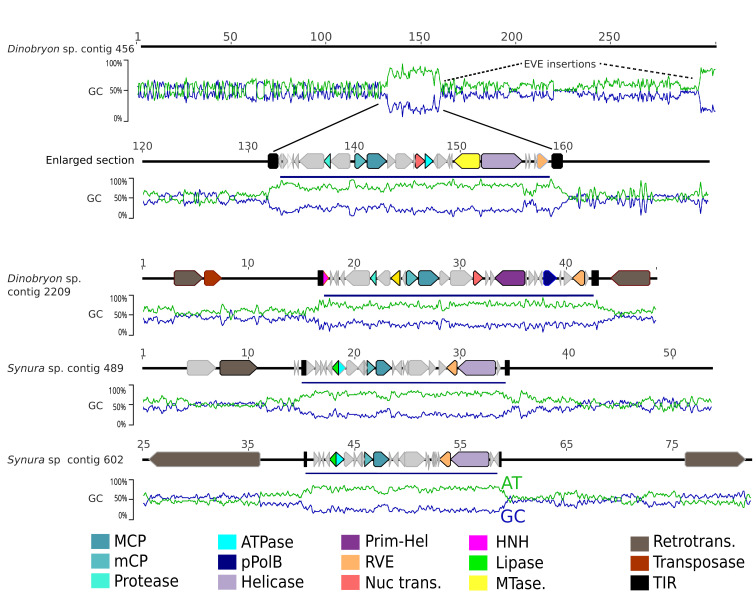
Fig. 3.
Protist genome assemblies from long-read data resolve full-length EVE insertions. Selected contigs from a partial assembly of Oxford Nanopore reads from Synura sp. LO234KE and Dinobryon sp. LO226KS showing virus insertion regions flanked by TIRs. Integrated viral regions often exhibit a notable difference in GC content from the host genome. Synura sp. contains Polinton-like EVEs, whereas Dinobryon sp. EVEs belong to the virophage group. Contig 456 features two insertions at positions 150 and 300 kbp, the latter being only partially assembled. Retrotransposons (Ty3/Gypsy; brown annotation) are often observed near virus insertions. Graphs below contigs show GC (blue) and AT (green) percentages. Numbers denote contig position (kbp). MCP – major capsid protein, mCP – minor capsid protein, Protease – cysteine protease, Helicase – DNA helicase, pPolB – DNA polymerase type B, Pri–Hel – Primase–Helicase, RVE – retroelement type integrase, Nuc trans. – nucleotidyl transferase, MTase – DNA methyltransferase, Retrotrans. – retrotransposon, TIR – terminal inverted repeat, HNH – homing endonuclease.