Significance
The Cuvierian organ (CO), first described by the French zoologist Georges Cuvier (1831), serves as a defensive organ with bioadhesive properties that is sacrificially ejected in some sea cucumber species. The genome of Holothuria leucospilota, an ecologically significant tropical sea cucumber with prototypical CO, unravels the molecular underpinnings of CO defense, which includes characteristic long-repeat signatures in CO-specific proteins enriched with amyloid-patterns. Genomic evidence further reveals that H. leucospilota directly transduces predator-triggered mechanical pressure onto the CO surface, leading to CO expulsion mediated by transient receptor potential channels and acetylcholine. The present study thus offers new insights into the molecular underpinnings of this unique organ in holothurian species.
Keywords: genome, tropical sea cucumber, bioadhesives, amyloid, transient receptor potential cation
Abstract
Some tropical sea cucumbers of the family Holothuriidae can efficiently repel or even fatally ensnare predators by sacrificially ejecting a bioadhesive matrix termed the Cuvierian organ (CO), so named by the French zoologist Georges Cuvier who first described it in 1831. Still, the precise mechanisms for how adhesiveness genetically arose in CO and how sea cucumbers perceive and transduce danger signals for CO expulsion during defense have remained unclear. Here, we report the first high-quality, chromosome-level genome assembly of Holothuria leucospilota, an ecologically significant sea cucumber with prototypical CO. The H. leucospilota genome reveals characteristic long-repeat signatures in CO-specific outer-layer proteins, analogous to fibrous proteins of disparate species origins, including spider spidroin and silkworm fibroin. Intriguingly, several CO-specific proteins occur with amyloid-like patterns featuring extensive intramolecular cross-β structures readily stainable by amyloid indicator dyes. Distinct proteins within the CO connective tissue and outer surface cooperate to give the expelled matrix its apparent tenacity and adhesiveness, respectively. Genomic evidence offers further hints that H. leucospilota directly transduces predator-induced mechanical pressure onto the CO surface through mediation by transient receptor potential channels, which culminates in acetylcholine-triggered CO expulsion in part or in entirety. Evolutionarily, innovative events in two distinct regions of the H. leucospilota genome have apparently spurred CO’s differentiation from the respiratory tree to a lethal defensive organ against predators.
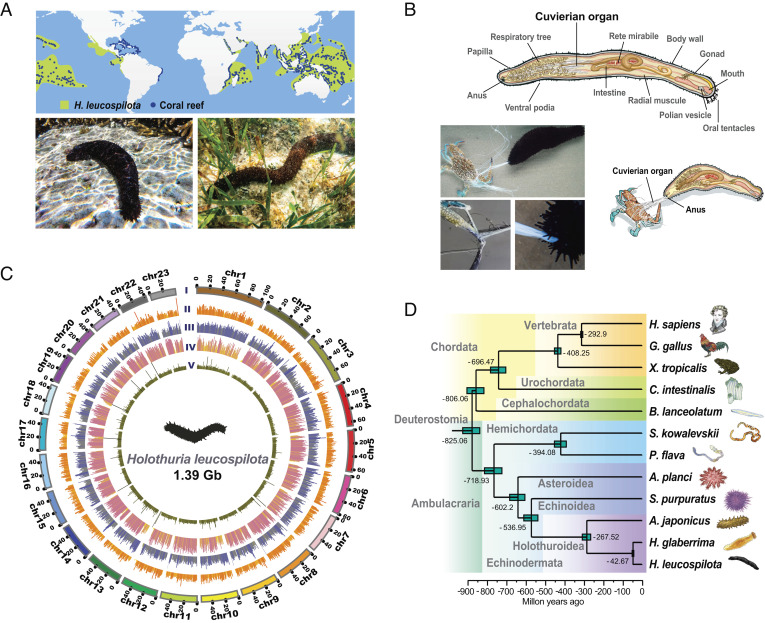
As a shallow-water holothurian, the tropical sea cucumber (Holothuria leucospilota) is widely distributed on a global scale. It thrives primarily in tropical localities of the Pacific and Indian Oceans (1), including some delicate marine habitats such as coral reefs (Fig. 1A) (2, 3). Despite its soft body and slow motion, H. leucospilota is surprisingly resilient against predation (4). To highlight, it leverages the Cuvierian organ (CO; also known as Cuvierian tubules as a tissue) for specialized defense (4). Named after Georges Cuvier, a French zoologist who first described it in 1831, the Cuvierian organ is a peculiar anatomical structure found in several species within the family Holothuriidae (5). As an internal organ embedded next to the gut, it branches off from the left respiratory tree and stays inactive in the posterior coelomic cavity (Fig. 1B) (6). When provoked by an offending mechanical stimulus, for example, tropical sea cucumbers respond by sacrificially ejecting the stored tubules through the cloacal orifice (anus), which then dramatically expand in length and adhesiveness within seconds (4). An ensnaring sticky matrix eventually immobilizes and traps potential predators, often to their demise (Movie S1) (7).
Fig. 1.
Geographical niches, genome landscape, and defense behavior of the tropical sea cucumber H. leucospilota. (A) Global distribution of H. leucospilota within or near coral reefs. (B) Schematic diagram showing the Cuvierian organ within the anatomy of H. leucospilota. The Cuvierian organ is located in the posterior coelomic cavity and is expelled toward a potential predator (crab). Details on Cuvierian tubules being ejected from the anus, with a crab being subsequently ensnared by Cuvierian tubules. (C) From outer to inner circles: CI, marker distribution across 23 chromosomes on megabase scales; CIIgene density; CIIIabundance of repetitive sequences; CIVSNP density and CVsequencing depth across the genome; CII–CV are drawn in non-overlapping 500-kb sliding windows. (D) Phylogeny of Deuterostomia, highlighting the position of H. leucospilota. Divergence times were estimated, as shown.
Remarkable anti-predation efficiency of the sea cucumber Cuvierian organ is apparently linked to markable tenacious and adhesive properties of its constituent molecules (Movie S2) (6, 8). During tubule elongation, externalization of granular cells in the outer layer of mesothelium occurs, which triggers granule release and rapid transformation of the Cuvierian organ into a sticky matrix upon encountering a foreign surface (9). The exact nature and operating principles of the tubules’ bioadhesive matrix remain poorly understood (7). In a similar vein, various forms of high-performance biological materials have been reported in animals, which exhibit either tenacity as seen in the silk fibers of spiders and silkworms (10, 11), or adhesiveness as seen in the byssus of mussels (12), cyprid footprint and cement of barnacles (13), and assemblies of other adhesive proteins in virtually unrelated species (14). With the advent of next-generation sequencing technologies, genomic approaches have proved useful in identification of functional components and design of biomimetic materials (11, 15).
The complete genomes of three echinoderm species, namely the purple sea urchin (Strongylocentrotus purpuratus) (16), crown-of-thorns starfish (Acanthaster planci) (17), and Japanese sea cucumber (Apostichopus japonicus, a member of the family Stichopodidae, which does not possess the Cuvierian organ) (18, 19), have been successfully sequenced, while low-coverage sequencing data have been made available for other species (20, 21). However, it remains quite challenging to deduce functional patterns from comparative genomics in marine invertebrates. This is because high diversity in morphology and physiology prevails between echinoderms and chordates and even between different classes of echinoderms at the genomic level, as previously illustrated by the complexity of innate immune recognition receptors (16, 22) and olfactory receptors (17).
Thus far, it remains unclear how adhesiveness genetically arose in the Cuvierian organ during its differentiation from the respiratory tree and how sea cucumbers perceive and transduce danger signals for Cuvierian organ expulsion in defense (4, 6). Here, we present a high-quality, chromosome-level genome assembly of H. leucospilota. Guided by these genomic resources, we demonstrate that proteins subserving adhesiveness and tenacity in the Cuvierian organ are mostly unique to H. leucospilota and that several Cuvierian organ-specific proteins with long-repeat signatures share a novel intramolecular cross-β organization like amyloid assembly. We further illustrate how H. leucospilota perceives the external pressure and switches on expulsion of the Cuvierian tubules by an acetylcholine signaling system. Collectively, this study provides the first genomic insights into defensive ensnarement in a representative species of marine deuterostome invertebrates.
Results
Genome Sequencing, Assembly, Annotation, and Phylogenetic Analysis.
Genomic DNA was isolated from a female individual of H. leucospilota and sequenced with 113.45-fold coverage of Illumina sequencing reads (Dataset S1 A) and 101.09-fold coverage of PacBio sequencing reads (Dataset S1 B). Based on these, a Hi-C library was constructed for chromosomal-level genome assembly (Dataset S1 C). On comparing several assemblers, we selected WTDGB for its advantages in continuity (Dataset S1 D). The final genome assembly is 1.39 Gb in size, with high heterozygosity (~2.11%) and multiplicity (~61.45%) (SI Appendix, Fig. S1). It encompasses 2,312 scaffolds with contig N50 and scaffold N50 sizes of 55.5 Mb and 56.1 Mb (Dataset S1 E), respectively, and features 97.63% high-quality read alignments (Dataset S1 F) and 96.3% BUSCO completeness (Dataset S1 G), indicating a significant improvement to all previously reported echinoderm genomes (contig N50: 0.01 to 0.19 Mb; scaffold N50: 0.07 to 1.52 Mb) (16–19). About 94.44% of the genome resides on the largest 23 scaffolds, which most likely correspond to the 23 chromosomes in the H. leucospilota genome (Fig. 1C, and SI Appendix, Fig. S2 and Dataset S1 H).
Genome annotation was approached by a combination of homologous comparison, Ribonucleic acid sequencing (RNAseq) prediction and ab initio prediction (Dataset S1 I). The H. leucospilota genome was predicted to contain 36,089 protein-coding genes (Dataset S1 J), of which 75.24% were annotated based on known protein public databases (SI Appendix, Fig. S3 and Dataset S1 K). The repeat content and transposable elements (TEs) account for 50.41% of the assembly (Dataset S1 L).
Phylogenetic analysis positioned H. leucospilota with A. japonicus and H. glaberrima within the class Holothuroidea, which is close to the class Echinoidea and the class Asteroidea (Fig. 1D and SI Appendix, Fig. S4). Diversification times of the families Stichopodidae and Holothuriidae in the order Aspidochirotida are estimated to be ~270 Mya, which are equivalent to diversification times of their vertebrate counterparts at the level of classes (Mammalia and Aves). This implies that differentiation among echinoderm families occurred as long-term and large-scale processes.
Structural Basis of the Cuvierian Tubules.
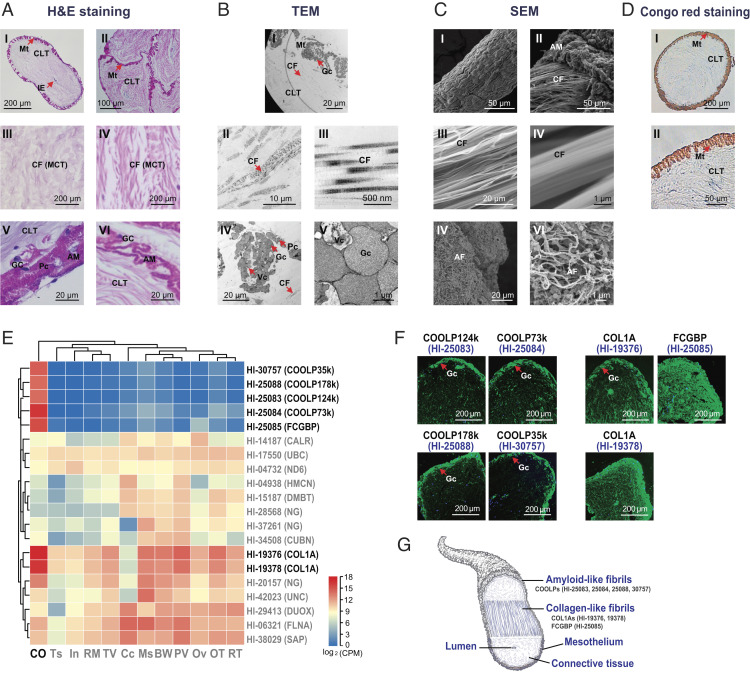
When encroached by a crab, for example, H. leucospilota veered its aboral end toward the source of assaults and underwent a whole-body contraction. The free ends of the Cuvierian organ were discharged through the anus to bind and entangle the crab, which was unable to break free upon ensnarement (Fig. 1B and Movie S1). When the microstructures were viewed in coronal sections by optical microscopy and transmission electron microscopy (TEM), the Cuvierian tubules of H. leucospilota were found to consist of an inner epithelium surrounding a narrow lumen (Fig. 2A), a thick connective tissue layer rich in mutable collagenous structures (Fig. 2 A and B), and a mesothelium lining the tubule surface decorated with adhesive molecules in granular cells (Fig. 2 A and B). Furthermore, two distant fibrils were observed inside and on the surface of the Cuvierian tubules by scanning electron microscopy (SEM), respectively (Fig. 2C). Of these, the collagen-like fibrils are located in the middle connective tissue layer and the deduced amyloid-like fibrils are located in the outer mesothelium layer. The latter were stainable by the amyloid indicator Congo red (Fig. 2 C and D).
Fig. 2.
(A) Microstructures as revealed by H&E staining. AI: Single Cuvierian tubules; AII: Adhesive Cuvierian tubules; middle layer with mutable collagenous tissues in single (AIII) and adhesive (AIV) Cuvierian tubules; outer-layer granular cells in single (AV) and adhesive (AVI) Cuvierian tubules. (B) Ultrastructures by TEM. BI: Overall appearance of a coronal section; BII–BIII: Middle-layer collagen-like fibrils; BIV–BV: Outer-layer granular cells. (C) Ultrastructures as revealed by SEM. Surface (CI) and inner (CII) of a Cuvierian tubule; BII–BIII: Middle-layer collagen-like fibrils; BIV–BV: Outer-layer amyloid fibrils. (D) Congo red-stained amyloid fibrils. Mt, mesothelium; CLT, connective tissue layer; IE, inner epithelium; Gc, granular cells; Pc, peritoneocytes; Vc, vacuole; MCT, mutable collagenous tissues; AM, adhesive materials; CF, collagen-like fibrils; AF: Amyloid-like fibrils. (E) Heatmap illustrating the most abundantly expressed genes with tandem repeats among different tissues. Full details on genes with tandem repeats in H. leucospilota and their expression patterns in different tissues are provided in Datasets S1 Q and S1 R. CO, Cuvierian organ; TS, testis; In, intestine; RM, rete mirabile; TV, transverse vessel; Cc, coelomocytes; Ms, muscle; BW, body wall; polian vesicle, PV; Ov, ovary; OT, oral tentacles; RT, respiratory tree. (F) Localization of putative CO-specific adhesive and reinforced proteins by immunofluorescence, including Hl-25083, Hl-25084, Hl-25088, Hl-30757, Hl-19376, Hl-19378, and Hl-25085. (G) Schematic diagram illustrating components of the Cuvierian tubules subserving tenacity and adhesion.
Expression of Outer-Layer Proteins Specific to the Cuvierian Organ.
For gene expression analysis, transcript levels were examined by mRNA sequencing (SI Appendix, Fig. S5 and Dataset S1 M). As defined by counts per million mapped reads (CPM), 611 tissue highly (CPMCO > 100; Dataset S1 N) and 198 tissue specifically (CPMCO >20 folds of median CPM; Dataset S1 O) expressed genes in the Cuvierian organ were identified, of which 69 (34.8%) of the Cuvierian organ specifically expressed genes matched the Cuvierian organ highly expressed genes (SI Appendix, Fig. S6 and Dataset S1 P). On the whole, these highly and specifically expressed genes were functionally implicated in extracellular matrix, stickiness, and neuroendocrine receptors (SI Appendix, Fig. S7).
A total of 2,161 tandem repeats, satisfying the criterion of having six repeats of no less than three amino acids in repeat length, were found in 1,488 genes from the H. leucospilota genome (SI Appendix, Fig. S8 and Dataset S1 Q). Transcript profiles in tissue distribution analysis reveal that the Cuvierian organ was the tissue most enriched with mRNA expression of tandem repeats containing genes (Dataset S1 R). Of the 20 most prolifically expressed tandem repeats contained genes, 18 were tissue highly expressed, and seven were tissue specifically expressed in the Cuvierian organ (Fig. 2E).
In immunofluorescence staining, Hl-25083, Hl-25084, Hl-25088, and Hl-30757 were located in the outer mesothelium layer of the Cuvierian tubules (Fig. 2F), and were accordingly named as Cuvierian organ outer-layer proteins (COOLPs). On the other hand, Hl-19376, Hl-19378, and Hl-25085 did not show such restricted localization (Fig. 2F).
Cuvierian Organ Outer-Layer Proteins Feature Long Tandem Repeats Signatures and Amyloid-pattern.
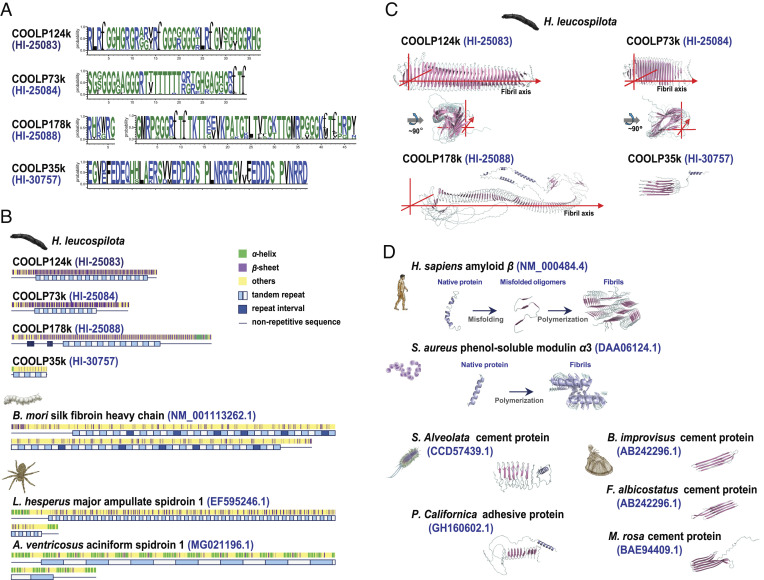
Interestingly, among the aforementioned seven CO-specific tandem repeats containing proteins, the four COOLPs (Hl-25083, Hl-25084, Hl-25088, and Hl-30757) have neither orthologues in other species (Dataset S1 S), nor known functional domains in their amino acid sequences, as revealed by annotation (SI Appendix, Fig. S9 and Dataset S1 T). Yet, they likewise possess high percentages (55.9 to 76.9%) of tandem repeats with long repeats (33 to 46 amino acids) (Fig. 3 A and B), suggesting the existence of a common mechanism afforded by such structural patterns. In contrast, the tandem repeats covered only a small part (0.6 to 1.4%) of the total amino acid sequences of Hl-19376, Hl-19378 and Hl-25085 (SI Appendix, Fig. S9). The repeat units seen in H. leucospilota CO-specific tandem repeats containing proteins turn out to be unique, with no sequence similarities with any known repeat motifs (Fig. 3A and SI Appendix, Fig. S9). Although tandem repeats have previously been observed in fibrous proteins in other species [such as spider spidroin (11) and silkworm fibroin (23)], the H. leucospilota COOLPs Hl-25083, Hl-25084, and Hl-25088 are arranged as full intramolecular β-sheets in their secondary structures, which are unprecedented in any fibrous proteins (Fig. 3B).
Fig. 3.
Amyloid-patterned Cuvierian organ outer-layer proteins (COOLPs) with long tandem repeats signatures. (A) Repeat units in H. leucospilota COOLPs Hl-25083, Hl-25084, Hl-25088, and Hl-30757. (B) Secondary structures, repeat lengths, and repeat numbers of COOLPs Hl-25083, Hl-25084, Hl-25088, and Hl-30757, as analyzed in a comparative perspective with silkworm fiborin (BM_001113262.1) and spider spidroins (EF595246.1 and MG021196.1). (C) AlphaFold-generated three-dimensional structures of the amyloid-patterned proteins COOLPs Hl-25083, Hl-25084, Hl-25088, and Hl-30757 in H. leucospilota. (D) Three-dimensional structures of other pathogenic and functional amyloids including human amyloid-beta (classical cross-β architecture), S. aureus PSMa3 (cross-α architecture), and adhesive or cement proteins from polychaete S. alveolate and P. californica, and barnacles B. improvises, F. albicostatus, and M. rosa.
The protein structures for H. leucospilota tandem repeats containing COOLPs and adhesive proteins in other species were predicted with AlphaFold (24) (SI Appendix, Fig. S10). Intriguingly, β-sheets are extensively present in Hl-25083, Hl-25084, and Hl-25088 which are serially arranged in an antiparallel manner to form cross-β structures that lie perpendicular to the peptide chain direction without polymerization (Fig. 3C). Thus, these proteins seem to be structurally defined by a novel amyloid architecture even as monomers, in comparison with known forms of amyloids including the canonical cross-β [as in human amyloid-beta (Ab) (25)], and cross-α [as in Staphylococcus aureus phenol-soluble modulin α3 (PSMa3) (26)], which are polymers from monomer aggregation (Fig. 3D). In addition, Hl-30757 does take the form of oligo-β-sheets similar to those reported for adhesive or cement proteins in polychaetes (14) and barnacles (27) (Fig. 3 C and D).
Pressure Induction leading to Cuvierian Organ Expulsion.
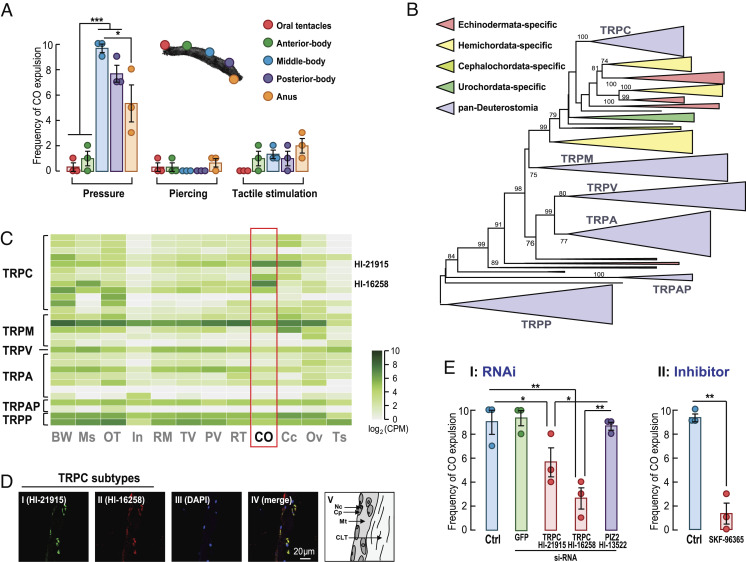
To stimulate Cuvierian organ expulsion in vivo, three methods of pressure application were used in five parts of H. leucospilota, namely the oral tentacles, anterior-, middle-, and posterior-bodies, and anus (Fig. 4A). Squeezing the internal organs to exert physical pressure on the middle- and posterior-bodies was the most effective way to trigger Cuvierian organ expulsion. On the other hand, sustained tactile stimulation on the posterior-body and anus could only occasionally induce expulsion, whereas piercing the skin with a needle failed to work in any parts of the sea cucumber (Fig. 4A).
Fig. 4.
Pressure sensing governs H. leucospilota Cuvierian organ expulsion. (A) of Cuvierian organ expulsion upon three different modes of stimulation at five different parts of the organism. The stimulation methods were pressure by squeezing, piercing, and tactile stimulation. The stimulated parts included the oral tentacles, anterior, middle-, and posterior-bodies, and anus. (B) Phylogenetic tree of the transient receptor potential channel genes in H. leucospilota and other deuterostome animals. A detailed version of the tree is provided in Dataset S1. (C) Heatmap illustrating the tissue distribution of TRP genes in H. leucospilota. (D) Fluorescence in situ hybridization (FISH) of TRPCs Hl-21915 and Hl-16258 mNRA in the Cuvierian tubules. (E) Involvement of TRPCs on Cuvierian organ expulsion induced by pressure. EI: Effects of RNAi targeting the TRPCs Hl-21915 and Hl-16258 and PIZ2 Hl-13522 on CO expulsion; EII: Effects of TRPC signaling blockage by SKF-96365 on CO expulsion. Behavioral data presented are expressed as mean ± SE (n = 3 individual groups, each of which containing 10 biological replicates). *P <0.05 and **P <0.01.
Mediation of Cuvierian Organ Expulsion by TRP Canonical (TRPCs).
As two transient receptor potential (TRP) channel genes (Hl-21915 and Hl-16258) are highly expressed in the Cuvierian organ (SI Appendix, Fig. S6 and Dataset S1 P), we set out to analyze tissue expression profiles of all members of the TRP superfamily in H. leucospilota. In conjunction with phylogenetic analysis on TRP superfamily genes in Deuterostomia (Fig. 4B, and Dataset S1 U and Dataset S2), it was confirmed that these two CO-specifically expressed TRP genes belong to TRP canonical (TRPC) channels (Fig. 4C). Fluorescence in situ hybridization showed that Hl-21915 and Hl-16258 mRNA co-localized in the outer mesothelium of Cuvierian tubules (Fig. 4D).
In RNA interference experiments, silencing of both TRPCs, Hl-21915, and Hl-16258, but not Hl-13522 (corresponding to Piezo-type mechanosensitive ion channel component 2; PIZ2), partially reduced expulsion behaviors under pressure (Fig. 4E). Likewise, blockage of the TRPC mediated Ca2+ entry was effective in inhibiting pressure-induced CO expulsion (Fig. 4E). Experimental results for gene knockdown and signaling blockage support the involvement of TRPCs in this sea cucumber-specific defensive behavior.
Neurotransmission Effects of Ach/nAchR on Cuvierian Organ Expulsion.
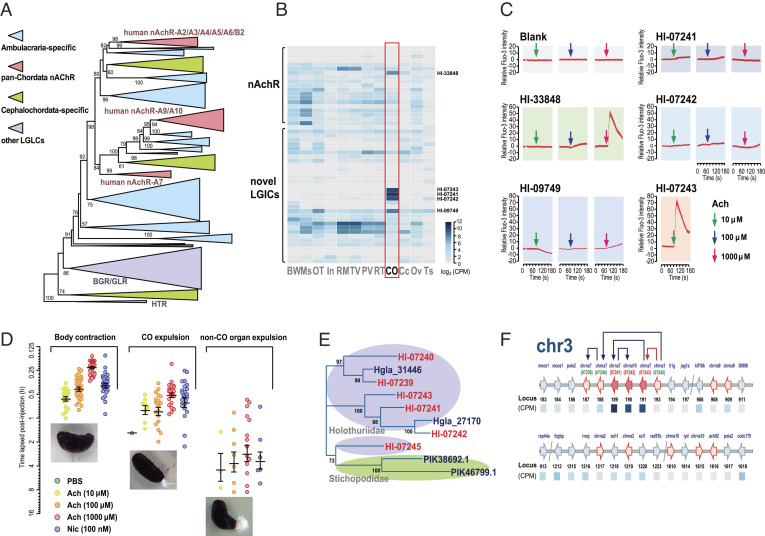
The ligand-gated ion channel (LGIC) superfamily emerged as another highly expressed gene family in the Cuvierian organ of H. leucospilota (SI Appendix, Fig. S7 and Dataset S1 P). By phylogenetic analysis on LGIC superfamily genes in Deuterostomia, a highly expanded group of Ambulacraria-specific genes was identified in addition to the typical nicotinic acetylcholine receptor (nAchR) genes (Fig. 5A and Dataset S1 V and Dataset S3). Among the novel Ambulacraria-specific LGIC genes, three (HL-07241, HL-07242, HL-07243) had the highest levels of tissue-specific expression in the Cuvierian organ (Fig. 5B). In addition, two other LGICs were highly and specifically expressed in the Cuvierian organ, within which Hl-33848 is a definite echinoderm nAchR, whereas Hl-09749 is relatively distant from the known nAchRs, based on phylogenetic classification (Fig. 5 A and B).
Fig. 5.
Roles of acetylcholine signal transduction in H. leucospilota Cuvierian organ expulsion. (A) Phylogenetic tree of ligand-gated ion channel superfamily genes in H. leucospilota and other deuterostome animals. A detailed version of the tree is provided in Dataset S2. (B) Heatmap illustrating the tissue distribution of nAchR-like genes in H. leucospilota. (C) Functional coupling of acetylcholine and putative Cuvierian tubule specifically expressed LGICs (Hl-07241, Hl-07242, Hl-07243, Hl-09749, Hl-33848) in Ca2+ mobilization. (D) Pharmacological effects of acetylcholine and nicotine on the behaviors of body contraction, Cuvierian organ expulsion, and non-CO organ expulsion in H. leucospilota. Behavioral data presented are expressed as mean ± SE (n = 30 individuals). (E) Phylogenetic analysis on H. leucospilota CO-specifically expressed nAchR-like LGIC genes, as viewed within Holothuroidea. (F) A model for chromosomal organization and expansion of CO-specific expressed nAchR-like LGIC genes, as viewed within the H. leucospilota chromosome 3.
Exogenously administered acetylcholine potently triggered a surge in intracellular free Ca2+ levels in HEK293 cells solely overexpressing H. leucospilota Hl-07243 at a low dose (10 μM), but not in cells overexpressing Hl-07241, Hl-07242 or Hl-09749 even at a high dose (1,000 μM) (Fig. 5C), suggesting a novel group of Ambulacraria-specific nAchRs being at work. In contrast, Ca2+ responses were triggered by acetylcholine on Hl-33848 (a typical nAchR) at a high dose (1,000 μM) (Fig. 5C).
In vivo administration of acetylcholine into the coelom of H. leucospilota induced ejection of the Cuvierian organ in time- and dose-dependent manners, which could be mimicked by nicotine, an exogenous ligand of nAchR (Fig. 5F). Additionally, two other aspects of motor behavior were collaterally stimulated by acetylcholine/nicotine injection. Body contraction preceded Cuvierian organ expulsion after, while non-CO organ expulsion only took place in a fraction (10.0 to 46.7%) of the tested individuals, following Cuvierian organ expulsion (Fig. 5D).
Within the novel group of Ambulacraria-specific nAchRs, the Cuvierian organ specifically expressed HL-07241, HL-07242, and HL-07243 were clustered into a subgroup present exclusively in the family Holothuriidae but not the family Stichopodidae (Fig. 5E). Based on evidence from synteny analysis, it could be logically deduced that Hl-07245 is an ancient gene in the class Holothuroidea, and that Hl-07243, Hl-07242, and Hl-07241 were generated by gene duplication in a sequential manner (Fig. 5F). Evidently, duplication of Hl-07243 from Hl-07245 was a key event enabling Cuvierian tubules to acquire the capability of controlled expulsion in sea cucumbers.
Discussion
Echinoderms are ancient marine invertebrates of the superphylum deuterostomes, comprising at least 13,000 fossil species and 7,000 living species (28). Sea cucumbers are representatives of the class Holothuroidea, form a large group (>1,693 species) with significant global distribution (29). Based on our genome-scale phylogenetic analysis, the divergence time between sea cucumber and sea urchin (~537 Mya) preceded the first appearance of sea cucumber fossil evidence in Middle Ordovician, or ~464 Mya (29). Thus, extreme evolutionary distances make it difficult to construe their specific life phenomena by comparison with mammals or other vertebrates, while genome-wide analysis serves as an attractive alternative (16).
Structural materials of living organisms generally evolve into functional maturity after long periods of selective processes and have served in many cases to inspire innovative materials design (10, 30, 31). The unique blend of strength and tenacity seen in some naturally occurring structural materials has often proved challenging to mimic synthetically (31). Examples include exceptionally tough insect silks (10), underwater adhesive mussel byssus (32), compression-resistant abalone nacre (33), and highly elastic insect resilin (34). In recent years, an impressive spectrum of proteins with novel or unknown functions have been unveiled by genomic sequencing of marine animals (15). This has gradually brought advances to our understanding of functionally versatile adhesive materials, though it remains at times difficult to isolate and pinpoint the exact proteins or molecules responsible for adhesion (35). Recently, AlphaFold has gained recognition as a powerful computational approach capable of accurately predicting protein structures with atomic precision even in cases where no similar structures have been reported (24). In this present study, the steady-state structures of H. leucospilota CO-specific proteins and adhesive proteins in other species can be relatively accurately predicted by AlphaFold (Fig. 3 C and D and SI Appendix, Fig. S10), though with prediction confidence levels lower than that for proteins from the human proteome (36).
In histological analyses on micro- and ultrastructures, collagen-like fibrils were found in the middle connective tissue layer (Fig. 2 B and C), whereas amyloid-like fibrils were observed in the outer mesothelium layer (Fig. 2 C and D), indicating that H. leucospilota Cuvierian tubules are compositionally defined by these two distinct types of fibrils in the thick middle layer for tenacity and the thin outer layer for adhesion, respectively (Fig. 2G). Importantly, the mutable collagenous tissue in the Cuvierian tubules is able to undergo rapid, though irreversible, changes in passive mechanical properties (Fig. 2A). The H. leucospilota CO-specific proteins containing long tandem repeats arranged in serial β-sheets (Fig. 3 A and B) are specifically located in the outer mesothelium layer (Fig. 2F). Extensive intramolecular serial β-sheets (n = 73 to 152 sheets) forming cross-β structures are a salient feature of some H. leucospilota CO-specific proteins (e.g., Hl-25083, Hl-25084, Hl-25088), which are not observed in known fibrous proteins, such as spider spidroin and silkworm fibroin (23) (Fig. 3B). Importantly, the H. leucospilota COOLPs share no sequence homology with any other marine adhesive proteins (Dataset S1 S). The sequence signatures that critically enable protein nanostructures to function as bioadhesives remain hitherto unelucidated (37), which is being first addressed in this study through analysis on three-dimensional (3D) structures of H. leucospilota CO-specific proteins (Fig. 3C and SI Appendix, Fig. S10).
Amyloid aggregates have been frequently implicated in the pathogenesis of various human diseases including Alzheimer’s disease, Parkinson's disease, type 2 diabetes, and prion diseases (38). However, some forms of amyloid fibrils occurring in bacteria, fungi, insects, and primates are known to perform physiological function in non-pathological contexts and are thus recognized as functional amyloids (39). The existence of amyloid-like fibrils on the surface of H. leucospilota Cuvierian tubules was strongly corroborated by evidence from ultrastructural imaging and histological staining with the amyloid indicator dye, Congo red (Fig. 2 C and D). For comparison, functional amyloids have been reported in bioadhesive materials formed by barnacle cement proteins, as revealed by amyloid dye staining (40).
In classical models of amyloid proteins including amyloid-β peptide (Aβ), islet amyloid polypeptide, and prion protein, formation of amyloid-like fibrils is associated with elevated β-structure content within the protein structure as a cross-β architecture, ultimately leading to its fibrillar aggregation (38) (Fig. 3D). Interestingly, protein assembly of PSMa3 in S. aureus was revealed to assume a novel cross-α amyloid-like architecture (26) (Fig. 3D). Here, our findings indicate that amyloid-like fibrils in H. leucospilota Cuvierian tubules arise possibly from proteins [COOLP124k (Hl-25083), COOLP73k (Hl-25084), and COOLP178k (Hl-25088)] containing serial oligo-β-sheets to constitute an intramolecular cross-β architecture (Fig. 3A), bearing resemblance to signatures of an amyloid assembly. On the other hand, the relatively small H. leucospilota COOLP35k (Hl-30757; 35 kDa) do contain a few oligo-β-sheets (n = 9 sheets), as in the case of barnacle cement proteins (19 kDa; n = 8 sheets) and polychaete adhesive proteins (22 to 27 kDa; n = 7 to 18 sheets) (Fig. 3 C and D), suggesting functional convergence via parallel mechanisms for bioadhesion among different species. Given that COOLPs Hl-25083, Hl-25084 and Hl-25088 co-localize in a 194-kb region in chromosome 12, we inferred that this region is a fundamental genetic tool kit involved in bioadhesion of the Cuvierian tubules.
It has been proposed that sea cucumbers perceive their predators, at least partially, by sensing hydrodynamic forces with their oral tentacles, tube feet, and spines (41). However, the exact mechanoreceptors involved have not been identified, with the exception of an ankyrin TRP responsive to flow velocity (42). Our current study shows that when physical pressure from squeezing was directly applied to the Cuvierian organ of H. leucospilota (Fig. 4A), the TRPCs (Hl-21915 and Hl-16258) located on the Cuvierian tubules outer mesothelium mediate this pressure (Fig. 4D), instead of PIZ2, a channel traditionally deemed sensitive to mechanical stimuli (Fig. 4E). This was experimentally demonstrated by both gene knockdown and signaling blockage. In contrast, piercing and tactile stimulation on the skin surface only weakly induced Cuvierian organ expulsion (Fig. 4A). This suggests that a stimulus-selective response in H. leucospilota may be turned on only when an encroaching predator exerts a sufficiently large physical force directly onto the Cuvierian organ to provoke bioadhesive ensnarement.
During self-defense by H. leucospilota, physical assaults from predators sensed by the TRPCs may be transduced into acetylcholine-triggered signals, which are eventually received by a novel nAchR (Hl-07243) specifically expressed in the Cuvierian organ (Fig. 5 B and C). Key evolutionary events for Hl-07243 duplication from Hl-07245 possibly allowed some tropical sea cucumbers in the family Holothuriidae to acquire the innovative capacity to expel Cuvierian tubules (Fig. 5 E and F), which was derived from the basal region of an otherwise non-defensive organ, the respiratory tree (6). Classically, acetylcholine is known to induce muscle shrinking (43) and gamete release (44) in sea cucumbers. Our study here has advanced the first mechanistic details on its regulatory roles in Cuvierian organ expulsion.
Taken as a whole, the complete genome of H. leucospilota, in conjunction with in vivo findings on transcriptional expression patterns and multiple behavioral traits analyzed in this study, offers fresh insights into the biological innovations and significance of the Cuvierian organ, a unique defense system engineered with long repeats containing amyloid-patterned proteins. While amyloids with cross-β structures have been implicated in the pathogenesis of various human diseases (45), their invertebrate counterparts, such as those illustrated here in H. leucospilota Cuvierian tubules and elsewhere as adhesive materials in other marine animals (40), can operate as pivotal components of physiological functions. Our study also illustrates the genomic basis for differentiation of the Cuvierian organ from the respiratory tree in the family Holothuriidae. The proteins identified containing long tandem repeats with serial β-sheets (Hl-25083, Hl-25084, and Hl-25088, Fig. 3C) suggest that the CO might have evolved adhesive properties through a separate outer layer of amyloid materials, while the nAchRs (Hl-07241, Hl-07242 and Hl-07243, Fig. 5F) provide a coordinated machinery guided by Ach signals for expelling Cuvierian tubules. Further investigation on whether amyloid-patterned proteins attain adhesiveness at the intramolecular and intermolecular levels should help advance our knowledge on non-pathological functions of amyloids in marine animals.
Materials and Methods
Further detailed information on the materials and methods used in this study can be found in the SI Appendix.
Sample Collection and Genome Sequencing.
The rete mirabile from a female adult H. leucospilota specimen was used for genome sequencing and construction of the Hi-C library. Short reads, long reads, and Hi-C sequencing were obtained on the HiSeq X Ten platform (Illumina), PacBio Sequel I system (PacBio), and BGI MGISEQ-2000 sequencer (BGI), respectively. All cleaned data were mapped onto contigs by using BWA aligner, while LACHESIS was used for scaffolds de novo assemblies. Chromosome-scale scaffolds were adjusted manually with the help of JucieBoxon, on the basis of an interaction map created by Juicer.
Genome Annotation and Gene Family Analyses.
Gene structures were predicted by homologous comparison, RNA-seq prediction, and ab initio prediction. Gene functions were annotated based on best-matched hits in SwissProt and Gene motifs and domains were identified by InterProScan. Proteomes for H. leucospilota, and other 12 genomes in typical deuterostome species were selected for gene family analyses. CPM of each gene was calculated. Cross-sample normalization was done by using DESeq2 based on the RNA sequencing of samples from 12 tissues, including the body wall, muscle, oral tentacles, intestine, rete mirabile, transverse vessel, polian vesicle, respiratory tree, Cuvierian organ, coelomocytes, ovary, and testis.
Histology.
Microstructures of Cuvierian tubules were stained with hematoxylin and eosin (H&E) in optical microscopy. CO paraffin sections were stained with Congo red dye for visualization of amyloid fibrils. Ultrastructures were viewed under an S-3400N scanning electron microscope (Hitachi) and an HT7800 transmission electron microscope (Hitachi). For fluorescence in situ hybridization, Hl-16258 and Hl-21915 antisense cRNA probes were labeled at the 5′-ends with the Cy5 and 6-FAM dyes, respectively. For immunofluorescence staining, CO paraffin sections were incubated with primary antibodies against Hl-25083, Hl-25084, Hl-25088, Hl-30757, Hl-19376, Hl-19378, and Hl-25085, respectively, followed by FITC labeled secondary antibody. Fluorescence images were acquired with a Digital Eclipse C1 Microscope (Nikon).
Cuvierian Organ Component Analyses.
High-expression genes in CO were defined with the following threshold: CPM of CO tubules 20 folds greater than the median CPM of 12 tissues. The tandem-repeated proteins were identified with Tandem Repeats Finder. 3D structures of the proteins were predicted by AlphaFold based on their amino acid sequences. Five models brought into the CASP14 and full databases were employed.
Gene Family Analyses.
TRP genes were selected based on the keyword “transient receptor potential” in the SwissProt database. LGIC genes thereof were selected based on the domains “IPR006029” and “IPR006202” in the InterProscan database. Multiple sequence alignments for all gene families mentioned above were built with the Multiple Alignment using Fast Fourier Transform (MAFFT) aligner, while their corresponding phylogeny was inferred with RAxML, followed by visualization with Evolview.
Receptor Function Analysis.
ORFs of potential nAchR receptor Hl-33848 and novel LGICs Hl-07241, Hl-07242, Hl-07243, and Hl-07249 were subcloned into pcDNA3.1/Zeo(−) and functionally expressed in HEK293T cells. Transfected cells were preloaded with the Ca2+-sensitive dye Fluo3/AM, while test substances were subsequently loaded. [Ca2+]i measurements at the single-cell level were analyzed in the TCS-SP5 Confocal System (Leica).
In Vivo Experiments.
Three methods, including physical pressure (by squeezing), piercing and tactile stimulation (by stroking) were applied to test for any stimulatory effects on CO expulsion on five stimulated sites of the sea cucumbers, namely the front end (oral tentacles), anterior-body, middle-body, posterior-body, and rear end (anus). CO expulsion behaviors were observed by visual inspection and recorded within 1 min after stimulation.
dsRNA interference was carried out to test the roles of TRPCs (Hl-16258 and Hl-21915) and PIEZ2 (Hl-13522) in CO expulsion in the sea cucumber, and EGFP was used as a targeting negative control. Inhibition of TRPC was achieved by celomic injection of its blocker SKF-96365. The effects of TRPCs on CO expulsion were evaluated following RNAi or signaling blockage. Pressure on the middle-body was applied to induce CO expulsion. Behaviors were observed by visual inspection and recorded photographically.
Pharmacological effects of acetylcholine and nicotine on CO expulsion and other behaviors were monitored following celomic injection. Behaviors such as body contraction, CO expulsion, and non-CO organ expulsion were recorded upon their appearance, 0 to 16 h after injection.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (PDF)
Dataset S03 (PDF)
H. leucospilota expel Cuvierian tubules toward a crab and the crab was tied up.
Biochemical properties as assessed in terms of adhesiveness and tenacity of H. leucospilota Cuvierian tubules.
Acknowledgments
This study was graciously supported by grants from the National Natural Science Foundation of China (42176132), the National Key R&D Program of China (2018YFD0901605 and 2020YFD0901100), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0402), the Key Deployment Project of Centre for Ocean Mega-Research of Science, the Chinese Academy of Sciences (COMS2020Q03), and Li Ka Shing Foundation (510858044).
Author contributions
T.C., C.R., N.-K.W., D.F., and C.H. designed research; T.C., A.Y., C.S., L.Z., Y.R., J.L., X.W., D.H., J.H., X.L., F.W., Z.E., C.C., and X.Z. performed research; T.C., C.R., P.L., X.J., Y.W., and C.H. contributed new reagents/analytic tools; T.C., C.R., N.-K.W., A.Y., C.S., D.F., and X.J. analyzed data; and T.C., C.R., N.-K.W., A.Y., C.S., D.F., and C.H. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Ting Chen, Email: chan1010@scsio.ac.cn.
Chaoqun Hu, Email: hucq@scsio.ac.cn.
Data, Materials, and Software Availability
The H. leucospilota genome project has been deposited in the DNA Data Bank of Japan/European Nucleotide Archive/GenBank (DDBJ/ENA/GenBank) under the accession JAIZAY000000000. The raw reads were submitted to the Sequence Read Archive (SRA) database under BioProject number PRJNA747844. The whole-genome sequencing data and RNA-seq data from various tissue transcriptomes were deposited with the SRA database under accession nos. SRR15244370–SRR15244388 and SRR15275174–SRR15275209, respectively (46).
Supporting Information
References
- 1.Purcell S. W., Samyn Y., Conand C., Commercially important sea cucumbers of the world. In: FAO Species Catalogue for Fishery Purposes. Rome 6, 67 (2012). [Google Scholar]
- 2.Schneider K., et al. , Potential influence of sea cucumbers on coral reef CaCO3 budget: A case study at One Tree Reef. J. Geophys. Res-Biogeo. 116, 04032 (2011). [Google Scholar]
- 3.Bonham K., Held E. E., Ecological observations on the sea cucumbers Holothuria atra and H. leucospilota at Rongelap Atoll, Marshall islands (1963).
- 4.Hamel J. F., Mercier A., Cuvierian tubules in tropical holothurians: Usefulness and efficiency as a defence mechanism. Mar. Freshw. Behav. Phys. 33, 115–139 (2000). [Google Scholar]
- 5.Cuvier G., Latreille P., “Georges Cuvier sea cucumber” in The Animal Kingdom Arranged in Conformity with Its Organization, Cuvier G., Carvill H., Eds. New York, 1831), vol. 2, p. 342. [Google Scholar]
- 6.Becker P. T., Flammang P., “Unravelling the sticky threads of sea cucumbers — a comparative study on Cuvierian tubule morphology and histochemistry” in Biological Adhesive Systems: From Nature to Technical and Medical Application, von Byern J., Grunwald I., Eds. (Springer Vienna, Vienna, 2010), pp. 87–98. [Google Scholar]
- 7.DeMoor S., Waite J. H., Jangoux M., Flammang P., Characterization of the adhesive from cuvierian tubules of the sea cucumber Holothuria forskali (Echinodermata, Holothuroidea). Mar. Biotechnol. (NY) 5, 45–57 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Zahn R. K., Müller W. E., Michaelis M., Sticking mechanisms in adhesive organs from a Holothuria. Res. Mol. Biol. 2, 47–88 (1973). [Google Scholar]
- 9.Vandenspiegel D., Jangoux M., Cuvierian tubules of the Holothuroid Holothuria forskali (Echinodermata): A morphofunctional study. Mar. Biol. 96, 263–275 (1987). [Google Scholar]
- 10.Omenetto F. G., Kaplan D. L., New opportunities for an ancient material. Science 329, 528–531 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babb P. L., et al. , The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat. Genet. 49, 895–903 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Priemel T., et al. , Microfluidic-like fabrication of metal ion-cured bioadhesives by mussels. Science 374, 206–211 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Gohad N. V., et al. , Synergistic roles for lipids and proteins in the permanent adhesive of barnacle larvae. Nat. Commun. 5, 4414 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Hennebert E., Maldonado B., Ladurner P., Flammang P., Santos R., Experimental strategies for the identification and characterization of adhesive proteins in animals: a review. Interface Focus 5, 20140064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerette P. A., et al. , Accelerating the design of biomimetic materials by integrating RNA-seq with proteomics and materials science. Nat. Biotechnol. 31, 908–915 (2013). [DOI] [PubMed] [Google Scholar]
- 16.S. U. G. S. Consortium et al. , The genome of the sea urchin Strongylocentrotus purpuratus. Science 314, 941–952 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall M. R., et al. , The crown-of-thorns starfish genome as a guide for biocontrol of this coral reef pest. Nature 544, 231–234 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zhang X. J., et al. , The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS. Biol. 15, e2003790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y. L., et al. , Sea cucumber genome provides insights into saponin biosynthesis and aestivation regulation. Cell Discov. 4, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long K. A., Nossa C. W., Sewell M. A., Putnam N. H., Ryan J. F., Low coverage sequencing of three echinoderm genomes: the brittle star Ophionereis fasciata, the sea star Patiriella regularis, and the sea cucumber Australostichopus mollis. Gigascience 5, 1–4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina-Feliciano J. G., Pirro S., Garcia-Arraras J. E., Mashanov V., Ryan J. F., Draft genome of the sea cucumber Holothuria glaberrima, a model for the study of regeneration. Front. Mar. Sci. 8, 603410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rast J. P., Smith L. C., Loza-Coll M., Hibino T., Litman G. W., Review - Genomic insights into the immune system of the sea urchin. Science 314, 952–956 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland C., Numata K., Rnjak-Kovacina J., Seib F. P., The biomedical use of silk: Past, present, future. Adv. Healthc. Mater. 8 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyama B. H., Weissman J. S., Amyloid structure: Conformational diversity and consequences. Annu. Rev. Biochem. 80, 557–585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tayeb-Fligelman E., et al. , The cytotoxic Staphylococcus aureus PSM alpha a3 reveals a cross-alpha amyloid-like fibril. Science 355, 831–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang C., et al. , Biochemistry of barnacle adhesion: An updated review. Front. Mar. Sci. 6, 565 (2019). [Google Scholar]
- 28.Bottjer D. J., Davidson E. H., Peterson K. J., Cameron R. A., Paleogenomics of echinoderms. Science 314, 956–960 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Miller A. K., et al. , Molecular phylogeny of extant Holothuroidea (Echinodermata). Mol. Phylogenet. Evol. 111, 110–131 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Bowen C. H., et al. , Microbial production of megadalton titin yields fibers with advantageous mechanical properties. Nat. Commun. 12, 5182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wegst U. G., Bai H., Saiz E., Tomsia A. P., Ritchie R. O., Bioinspired structural materials. Nat. Mater. 14, 23–36 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Priemel T., Degtyar E., Dean M. N., Harrington M. J., Rapid self-assembly of complex biomolecular architectures during mussel byssus biofabrication. Nat. Commun. 8, 14539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gim J., et al. , Nanoscale deformation mechanics reveal resilience in nacre of Pinna nobilis shell. Nat. Commun. 10, 4822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin G., Hu X., Cebe P., Kaplan D. L., Mechanism of resilin elasticity. Nat. Commun. 3, 1003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davey P. A., et al. , Omics-based molecular analyses of adhesion by aquatic invertebrates. Biol. Rev. Camb. Philos. Soc. 96, 1051–1075 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Tunyasuvunakool K., et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.So C. R., et al. , Sequence basis of barnacle cement nanostructure is defined by proteins with silk homology. Sci. Rep. 6, 36219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenberg D., Jucker M., The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fowler D. M., Koulov A. V., Balch W. E., Kelly J. W., Functional amyloid –from bacteria to humans. Trends Biochem. Sci. 32, 217–224 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Barlow D. E., et al. , Characterization of the adhesive plaque of the barnacle Balanus amphitrite: Amyloid-like nanofibrils are a major component. Langmuir 26, 6549–6556 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Brown N. A. W., Wilson D. R., Gagnon P., Plasticity in the antipredator behavior of the orange-footed sea cucumber under shifting hydrodynamic forces. Curr. Zool. 65, 685–695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin C., et al. , Effects of different flow velocities on behavior and TRPA1 expression in the sea cucumber Apostichopus japonicas. J. Oceanol. Limnol. 38, 1328–1340 (2020). [Google Scholar]
- 43.Welsh J. H., Marine invertebrate preparations useful in the bioassay of acetylcholine and 5-hydroxytryptamine. Nature 173, 955–956 (1954). [DOI] [PubMed] [Google Scholar]
- 44.Ikegami S., Kanatani H., Koide S. S., Gamete-release by 1-methyladenine in vitro in the sea cucumber, Leptosynapta inhaerens. Biol. Bull. 150, 402–410 (1976). [DOI] [PubMed] [Google Scholar]
- 45.Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Chen T., Holothuria leucospilota isolate:nanhai20183. NCBI BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA747844/. Deposited 18 July 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (PDF)
Dataset S03 (PDF)
H. leucospilota expel Cuvierian tubules toward a crab and the crab was tied up.
Biochemical properties as assessed in terms of adhesiveness and tenacity of H. leucospilota Cuvierian tubules.
Data Availability Statement
The H. leucospilota genome project has been deposited in the DNA Data Bank of Japan/European Nucleotide Archive/GenBank (DDBJ/ENA/GenBank) under the accession JAIZAY000000000. The raw reads were submitted to the Sequence Read Archive (SRA) database under BioProject number PRJNA747844. The whole-genome sequencing data and RNA-seq data from various tissue transcriptomes were deposited with the SRA database under accession nos. SRR15244370–SRR15244388 and SRR15275174–SRR15275209, respectively (46).