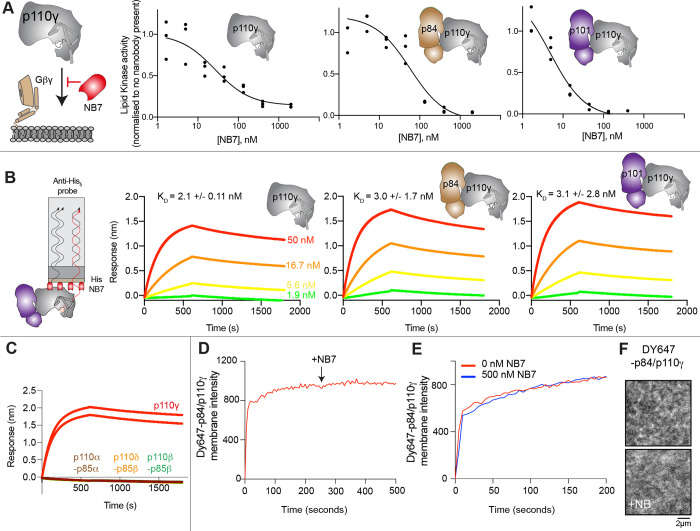
Figure 1. The inhibitory nanobody NB7 binds tightly to all p110γ complexes and inhibits kinase activity, but does not prevent membrane binding.
A. Cartoon schematic depicting nanobody inhibition of activation by lipidated Gβγ (1.5 μM final concentration) on 5% PIP2 membrane (5% phosphatidylinositol 4,5-bisphosphate (PIP2), 95% phosphatidylserine (PS)) activation. Lipid kinase assays showed a potent inhibition of lipid kinase activity with increasing concentrations of NB7 (3–3000 nM) for the different complexes. Experiments are carried out in triplicate (n=3) with each replicate shown. The y-axis shows lipid kinase activity normalised for each complex activated by Gβγ in the absence of nanobody. Concentrations of each protein were selected to give a lipid kinase value in the detectable range of the ATPase transcreener assay. The protein concentration of p110γ (300 nM), p110γ-p84 (330 nM) and p110γ-p101 (12 nM) was different due to intrinsic differences of each complex to be activated by lipidated Gβγ and is likely mainly dependent for the difference seen in NB7 response.
B. Association and dissociation curves for the dose response of His-NB7 binding to p110γ, p110γ-p84 and p110γ-p101 (50 – 1.9 nM) is shown. A cartoon schematic of BLI analysis of the binding of immobilized His-NB7 to p110γ is shown on the left. Dissociation constants (KD) were calculated based on a global fit to a 1:1 model for the top three concentrations and averaged with error shown. Error was calculated from the association and dissociation value (n=3) with standard deviation shown. Full details are present in the source data.
C. Association and dissociation curves for His-NB7 binding to p110γ, p110α-p85α, p110β-p85β, and p110δ-p85β. Experiments were performed in duplicate with a final concentration of 50 nM of each class I PI3K complex.
D. Effect of NB7 on PI3K recruitment to supported lipid bilayers containing H-Ras (GTP) and farnesyl-Gβγ as measured by Total Internal Reflection Fluorescence Microscopy (TIRF-M). DY647-p84/p110γ displays rapid equilibration kinetics and is insensitive to the addition of 500 nM nanobody (black arrow, 250 sec) on supported lipid bilayers containing H-Ras (GTP) and farnesyl-Gβγ.
E. Kinetics of 50 nM DY647-p84/p110γ membrane recruitment appears indistinguishable in the absence and presence of nanobody. Prior to sample injection, DY647-p84/p110γ was incubated for 10 minutes with 500 nM nanobody.
F. Representative TIRF-M images showing the localization of 50 nM DY647-p84/p110γ visualized in the absence or presence of 500 nM nanobody (+NB7). Membrane composition for panels C-E: 93% DOPC, 5% DOPS, 2% MCC-PE, Ras (GTP) covalently attached to MCC-PE, and 200 nM farnesyl-Gβγ.