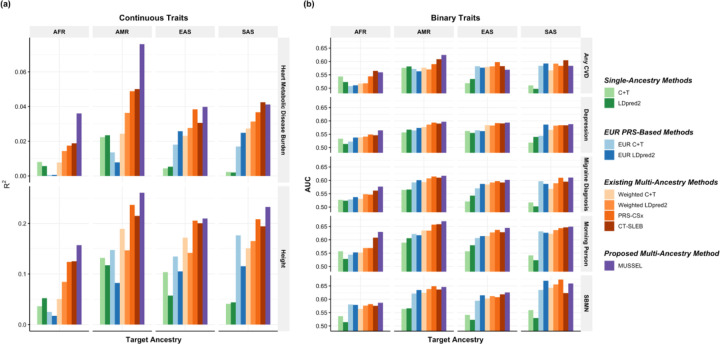
Figure 6: Prediction results on 23andMe validation individuals based on discovery GWAS from 23andMe on EUR, African American (AFR), Latino (AMR), EAS, and SAS.
The performance of the various methods is evaluated by (a) residual R2 for two continuous traits, heart Metabolic Disease Burden and height, and (b) residual AUC for five binary traits, any CVD, depression, migraine diagnosis, morning person, and SBMN. The LD reference data is from the 1000 Genomes Project (498 EUR, 659 AFR, 347 AMR, 503 EAS, 487 SAS). The dataset is randomly split into 70%, 20%, 10% for training GWAS, model tuning (tuning model parameters and training the SL in CT-SLEB and MUSSEL or the linear combination model in weighted LDpred2 and PRS-CSx), and testing (to report residual R2 or AUC values after adjusting for the top 5 genetic principal components, sex, and age), respectively. All methods were evaluated on the ~2.0 million SNPs that are available in HapMap3 + MEGA, except for PRS-CSx which is evaluated based on the HapMap 3 SNPs only, as implemented in their software. Ancestry- and trait-specific sample sizes of GWAS, number of SNPs included, and validation sample sizes are summarized in Supplementary Table 6.1.