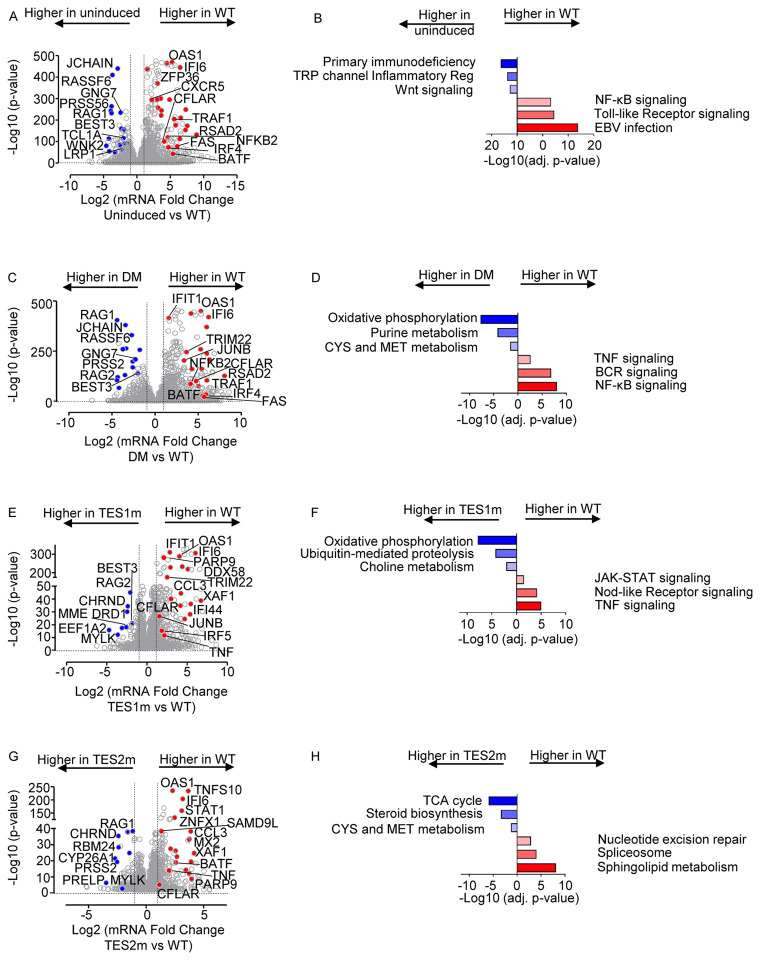
Fig. 6. Characterization of Akata B-cell pathways targeted by TES1 vs TES2 signaling.
(A) Volcano plot analysis of host transcriptome-wide genes differentially expressed in Akata cells conditionally induced for WT LMP1 expression for 24h by 250 ng/ml Dox versus in mock induced cells. Higher X-axis fold changes indicate genes more highly expressed in cells with WT LMP1 expression, whereas lower X-axis fold changes indicate higher expression in cells mock induced for LMP1. Data are from n=3 RNAseq datasets, as in Fig. 5.
(B) Enrichr analysis of KEGG pathways most highly enriched in RNAseq data as in (A) amongst genes more highly expressed in Akata with WT LMP1 (red) vs amongst genes more highly expressed with mock LMP1 induction (blue).
(C) Volcano plot analysis of host transcriptome-wide genes differentially expressed in Akata cells conditionally induced for WT vs DM LMP1 expression for 24h by 250 ng/ml Dox. Higher X-axis fold changes indicate genes more highly expressed in cells with WT LMP1 expression, whereas lower X-axis fold changes indicate higher expression in cells with DM LMP1. Data are from n=3 RNAseq datasets, as in Fig. 5.
(D) Enrichr analysis of KEGG pathways most highly enriched in RNAseq data as in (C) amongst genes more highly expressed in Akata with WT LMP1 (red) vs amongst genes more highly expressed with DM LMP1 induction (blue).
(E) Volcano plot analysis of host transcriptome-wide genes differentially expressed in Akata cells conditionally induced for WT vs TES1m LMP1 expression for 24h by 250 ng/ml Dox. Higher X-axis fold changes indicate genes more highly expressed in cells with WT LMP1 expression, whereas lower X-axis fold changes indicate higher expression in cells with TES1m LMP1. Data are from n=3 RNAseq datasets, as in Fig. 5.
(F) Enrichr analysis of KEGG pathways most highly enriched in RNAseq data as in (E) amongst genes more highly expressed in Akata with WT LMP1 (red) vs amongst genes more highly expressed with TES1m LMP1 induction (blue).
(G) Volcano plot analysis of host transcriptome-wide genes differentially expressed in Akata cells conditionally induced for WT vs TES2m LMP1 expression for 24h by 250 ng/ml Dox. Higher X-axis fold changes indicate genes more highly expressed in cells with WT LMP1 expression, whereas lower X-axis fold changes indicate higher expression in cells with TES2m LMP1. Data are from n=3 RNAseq datasets, as in Fig. 5.
(H) Enrichr analysis of KEGG pathways most highly enriched in RNAseq data as in (G) amongst genes more highly expressed in Akata with WT LMP1 (red) vs amongst genes more highly expressed with TES2m LMP1 induction (blue).