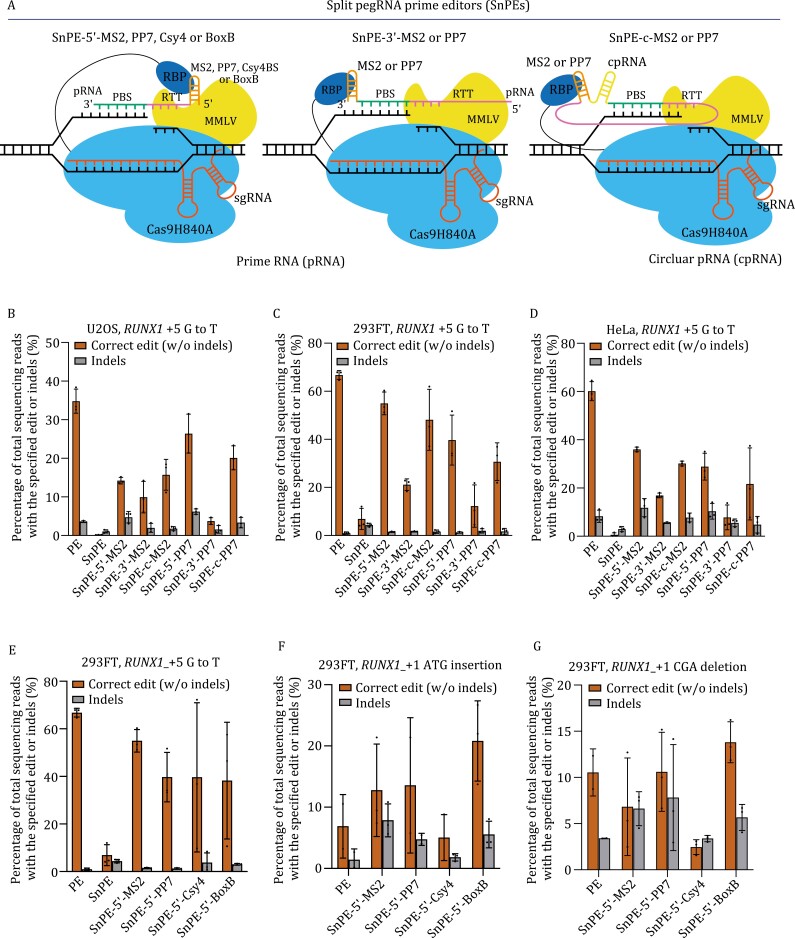
Figure 2.
Split pegRNA prime editing maintains PE activity by tethering prime RNA to Cas9. (A) Schematics of split pegRNA prime editors. The split pegRNA prime editor (SnPE) consists of a Cas9 nickase-H840A fused with C-terminal MMLV and N-terminal RNA binding proteins (RBPs), a sgRNA, and a separated prime RNA (pRNA). pRNA consists of a prime binding site (PBS), a reverse transcription template (RTT) and a RNA stem-loop aptamer such as MS2 or PP7. SnPE-5ʹ or 3ʹ-MS2 or PP7 consists of RBP-Cas9 nickase-MMLV, a pRNA bearing MS2 or PP7 and a sgRNA. RBP at the N-terminal Cas9 binds the RNA aptamer MS2 or PP7. SnPE-c-MS2 or PP7 includes the circular pRNA (cpRNA) bearing MS2 or PP7 generated by the tornado circular RNA expression system. Efficiency of SnPE-5ʹ-MS2 or PP7, SnPE-3ʹ-MS2 or PP7, SnPE-c-MS2, or PP7 were tested at RUNX1_+5 G·C to T·A using PE3 in U2OS cells (B), HEK293FT cells (C), and HeLa cells (D). (E) Efficiency of PE, SnPE, SnPE-5ʹ-Com, SnPE-5ʹ-BoxB, and SnPE-5ʹ-Csy4 at RUNX1_+5 G·C to T·A using PE3 in HEK293FT cells. (F) Efficiency of SnPE-5ʹ-MS2 or PP7, SnPE-5ʹ-MS2 or PP7, SnPE-5ʹ-Csy4 and SnPE-5ʹ-BoxB were tested at RUNX1_+1 ATG insertion using PE3 HEK293FT cells. (G) Efficiency of SnPE-5ʹ-MS2 or PP7, SnPE-5ʹ-MS2 or PP7, SnPE-5ʹ-Csy4, and SnPE-5ʹ-BoxB were tested at RUNX1_+1 CGA deletion using PE3 HEK293FT cells. Data and error bars in (B–G) indicate the mean and standard deviation of three independent biological replicates.