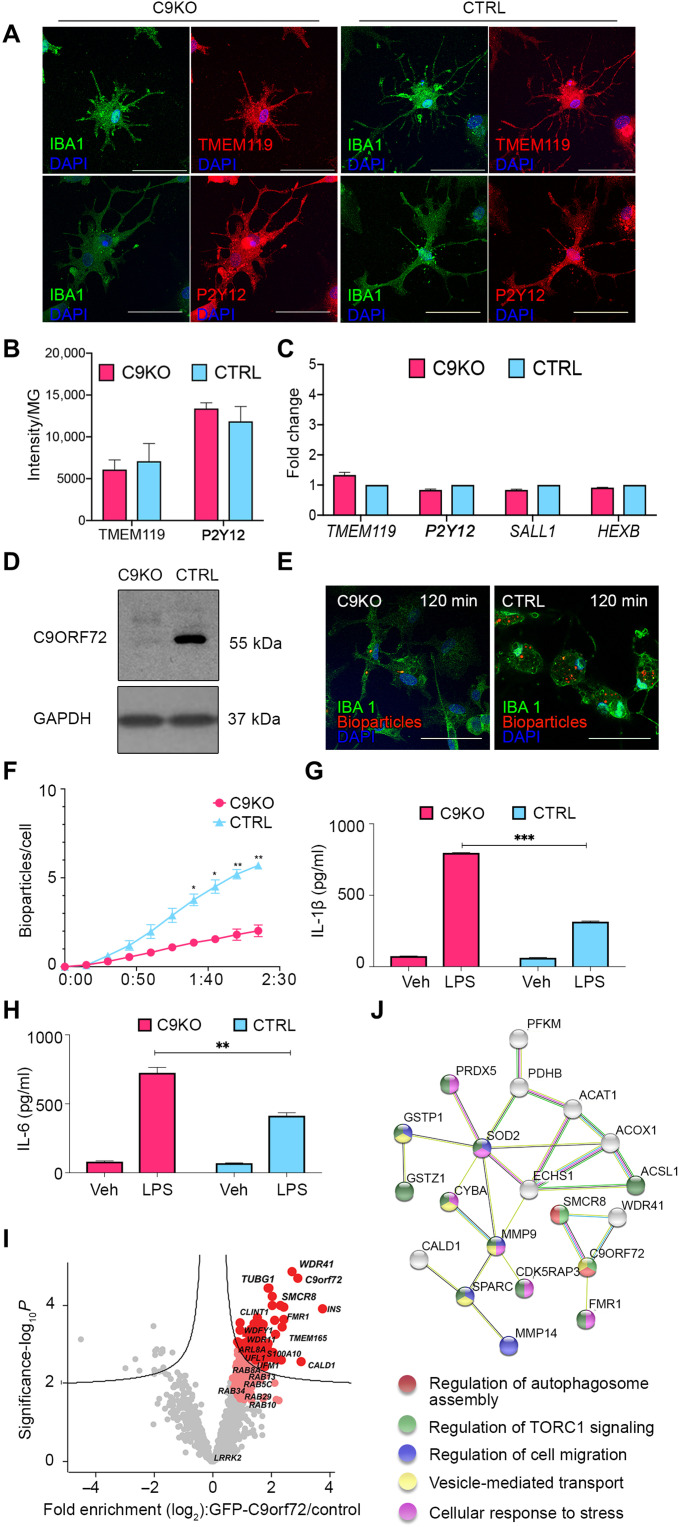
Fig. 3. C9KO-MG phenocopies functional deficits found in mC9-MG and mass spectrometric analysis of C9ORF72 interactome revealed its association with regulators of autophagy.
(A) Represents immune staining of C9KO-MG and CTRL-MG from the same genetic background confirming positive staining for microglial markers such as TMEM119 (red) and P2Y12 (red) along with IBA1 (green). Scale bars, 50 μm. (B) Bar graph representing equivalent fluorescence intensity per hiPSC-MG for TMEM119 and P2Y12 across C9KO-MG and CTRL-MG. (C) Bar graph representing the comparable expression of MG signature genes (TMEM119, P2Y12, SALL1, and HEXB) in C9KO-MG and CTRL-MG. Data are represented as means ± SD; N = 3. (D) Immunoblot validating loss of C9ORF72 protein (55 kDa) in C9KO-MG. (E) Representative images of immunofluorescence staining showing phagocytosis assay performed with pH-sensitive zymosan bioparticles for C9KO-MG and CTRL-MG at 120 min [IBA-1 (green), zymosan bioparticles (red)]. Scale bars, 50 μm. (F) Graph showing real-time imaging of zymosan bioparticle uptake at 15-min intervals, demonstrating a phagocytic deficit in C9KO-MG when compared to control (CTRL-MG). Statistical analysis was performed using two-way ANOVA and Tukey’s multiple comparisons test. Data are represented as means ± SEM (*P ≤ 0.05 and **P ≤ 0.01); N = 3. (G and H) Graphs demonstrating the increased production of IL-1β and IL-6 for C9KO-MGs following LPS stimulation; cells were treated with either LPS or 1× PBS [vehicle control (veh)]. Statistical analysis was performed using two-way ANOVA and Tukey’s multiple comparisons test. Data are represented as means ± SD; N = 3 (**P ≤ 0.01 and ***P ≤ 0.001). (G) Volcano plot demonstrating enrichment of C9ORF72 interactors at 1% false discovery rate (FDR) (dark red) and 5% FDR (light red). n = 4. (H) STRING network analysis of the C9ORF72 interactors (1% FDR); nodes represent the functional enrichment of the network. N for all experiments represents the number of times experiments were performed using cells generated from independent iPSC differentiations.