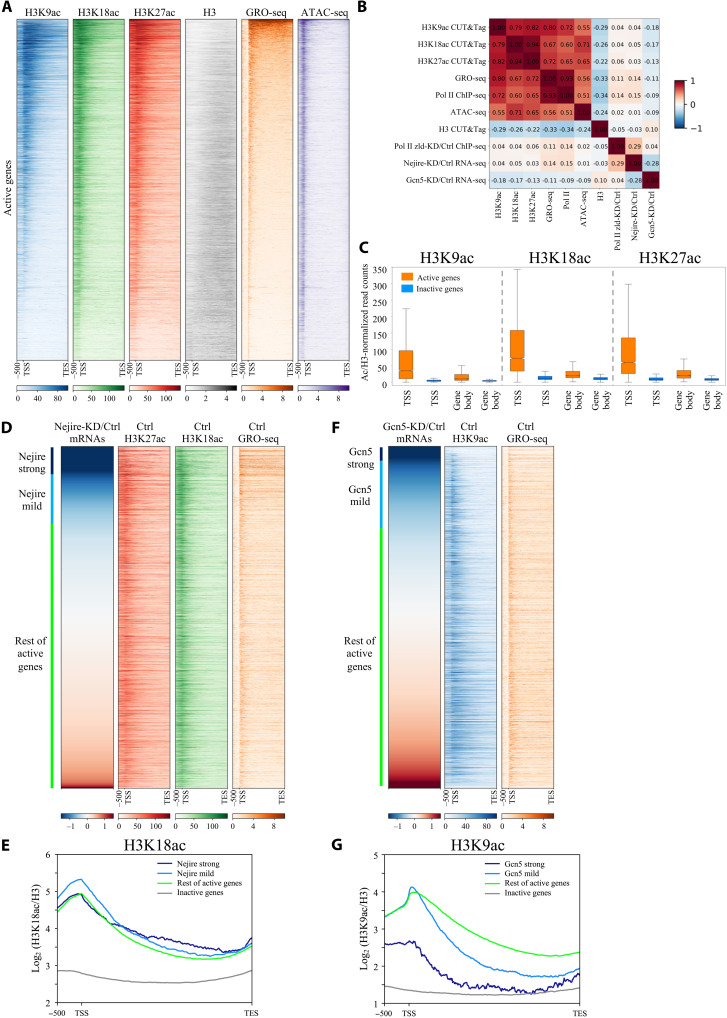
Fig. 4. Nejire and Gcn5 deposit their acetylation marks on every active gene, independently of their transcriptional coactivator activities.
(A) Heatmaps of ZGA active genes spanning from 500-bp upstream of TSS to TES. Genes are ranked by GRO-seq signal in column 5 (orange). Columns represent wild-type embryos H3K9ac CUT&Tag (blue), H3K18ac CUT&Tag (green), H3K27ac CUT&Tag (red), H3 CUT&Tag (gray), GRO-seq (orange) (15), and ATAC-seq (purple) (15) (n = 2 biological replicates). (B) Spearman’s correlation heatmap matrix comparing signal on gene body across multiple datasets on active genes in ZGA embryos. Numbers indicate R values. (C) Box plots comparing regions of active versus inactive genes at ZGA for H3K9ac, H3K18ac, and H3K27ac marks, normalized on H3. Boxes extend from 25th to 75th percentile of the data, and the whiskers span across the whole data range. TSS regions are defined as 400 bp surrounding TSS. Gene body regions are defined as 200-bp downstream of the TSS to TES. (D and F) Heatmaps of active genes at ZGA. Genes are ranked according to the ratio of RNA-seq levels of wild-type over Nejire-KD (D) or Gcn5-KD (F). Column color code corresponds to (A). Strong Nejire (D) or strong Gcn5 targets (F) (log2FC < −1.5) are marked in dark blue, mild Nejire (D) or mild Gcn5 targets (F) (−0.5 < log2FC < −1.5) are marked in light blue, and the rest of active genes are marked in green. (E and G) Metagene profiles of the H3K18ac signal (E) or H3K9ac (G) signal, normalized to H3, on strong Nejire (E) or strong Gcn5 (G) targets (dark blue), mild Nejire (E) or mild Gcn5 (G) targets (light blue), the rest of active genes (green), and inactive genes (orange) at ZGA. Numbers on y axis indicate the mean coverage of log2 (H3K18ac/H3) (E) or (H3K9ac/H3) (G).