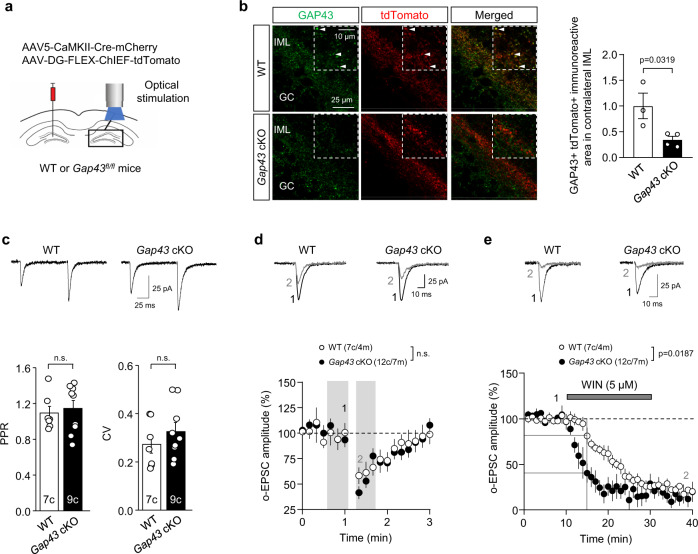
Fig. 5. GAP43 genetic deletion from MCs enhances CB1R function at MC-GC synapses.
a Schematic diagram illustrating the injection of a mix of AAV5-CamKII-Cre-mCherry and AAV-DG-FLEX-ChIEF-tdTomato in the hilus of 3-week-old Gap43fl/fl and WT mice. Light pulses of 0.5–2.0 ms were used to evoke EPSCs driven by the ChIEF-expressing axons originating from commissural MCs. b Reduced GAP43 immunoreactivity (green) in AAV-infected tdTomato + (red) fibers of the contralateral IML from injected Gap43 cKO mice compared to WT mice. Arrowheads point to colocalizing boutons in WT mice. Representative images and quantification of GAP43/tdTomato colocalization are shown (means ± SEM; WT n = 3 mice, Gap43 cKO n = 4 mice; two-tailed unpaired Student’s t test). c Whole-cell patch-clamp recordings of GCs were performed in Gap43 cKO and WT mice. Representative traces (top) and quantification bar graph for basal PPR and CV (bottom) are shown (means ± SEM; c = cells; n.s. by two-tailed unpaired Student’s t test). d Representative traces (top) and time-course summary plot (bottom) for DSE are shown (means ± SEM; c = cells, m = mice; shaded areas indicate the time intervals at which the statistical analysis was conducted; n.s. by two-tailed unpaired Student’s t test). e Representative traces (top) and time-course summary plot (bottom) for o-EPSC amplitude upon WIN-55,212-2 bath application (WIN; 5 μM, 20 min) (means ± SEM; c = cells, m = mice; WT vs Gap43 cKO at time15 min, p = 0.0187 by two-tailed unpaired Student’s t test). Source data are provided as a Source data file.