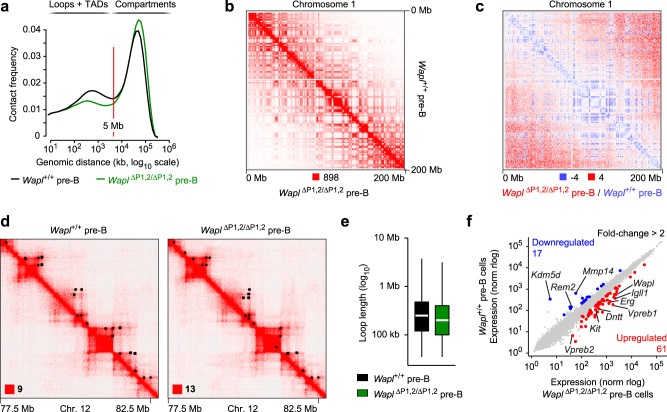
Fig. 2. Minor differences in the chromosomal architecture of Wapl+/+ and Wapl∆P1,2/∆P1,2 pre-B cells.
a Frequency distribution of intrachromosomal contacts as a function of the genomic distance using logarithmically increased genomic distance bins, determined by Hi-C analysis of ex vivo sorted Wapl+/+ (black) and Wapl∆P1,2/∆P1,2 (green) pre-B cells using HOMER (see Methods). b Hi-C contact matrices of chromosome 1, determined for ex vivo sorted Wapl+/+ and Wapl∆P1,2/∆P1,2 pre-B cells and plotted at a 500-kb bin resolution with Juicebox74. The intensity of each pixel represents the normalized number of contacts between a pair of loci5. The maximum pixel intensity is indicated below (red square). c Differential Hi-C contact matrix of chromosome 1 displaying the difference in pixel intensity between Wapl+/+ pre-B and Wapl∆P1,2/∆P1,2 pre-B cells as the ratio (Wapl∆P1,2/∆P1,2)/(Wapl+/+). More interactions (blue) in the TAD range were observed for Wapl+/+ pro-B cells, while more interactions (red) in the compartment range were detected for Wapl∆P1,2/∆P1,2 pre-B cells. d Hi-C contact matrices of a zoomed-in region on chromosome 12 (mm9; 77,500,000–82,500,000), shown for Wapl+/+ and Wapl∆P1,2/∆P1,2 pre-B cells. Black dots indicate loop anchors identified with Juicebox. The maximum pixel intensity is indicated in the lower left of the panel (red square). e Distribution of the loop length (in kb). White lines indicate the median and boxes represent the middle 50% of the data. Whiskers denote all values of the 1.5× interquartile range. The median loop length is 250 kb in Wapl+/+ pre-B cells (black) and 200 kb in Wapl∆P1,2/∆P1,2 pre-B cells (green). f Scatterplot of gene expression differences between Wapl∆P1,2/∆P1,2 and Wapl+/+ pre-B cells isolated from the bone marrow of the respective mice. Genes upregulated (red) or downregulated (blue) in Wapl∆P1,2/∆P1,2 pre-B cell compared with Wapl+/+ pre-B cells were analyzed with DESeq2 and defined by an expression difference of >2-fold, an adjusted P value of <0.05 and a TPM value of >5 in at least one of the two pre-B cell types (Supplementary Data 2). Two RNA-seq experiments were performed with ex vivo sorted pre-B cells of each genotype.