Figure 1.
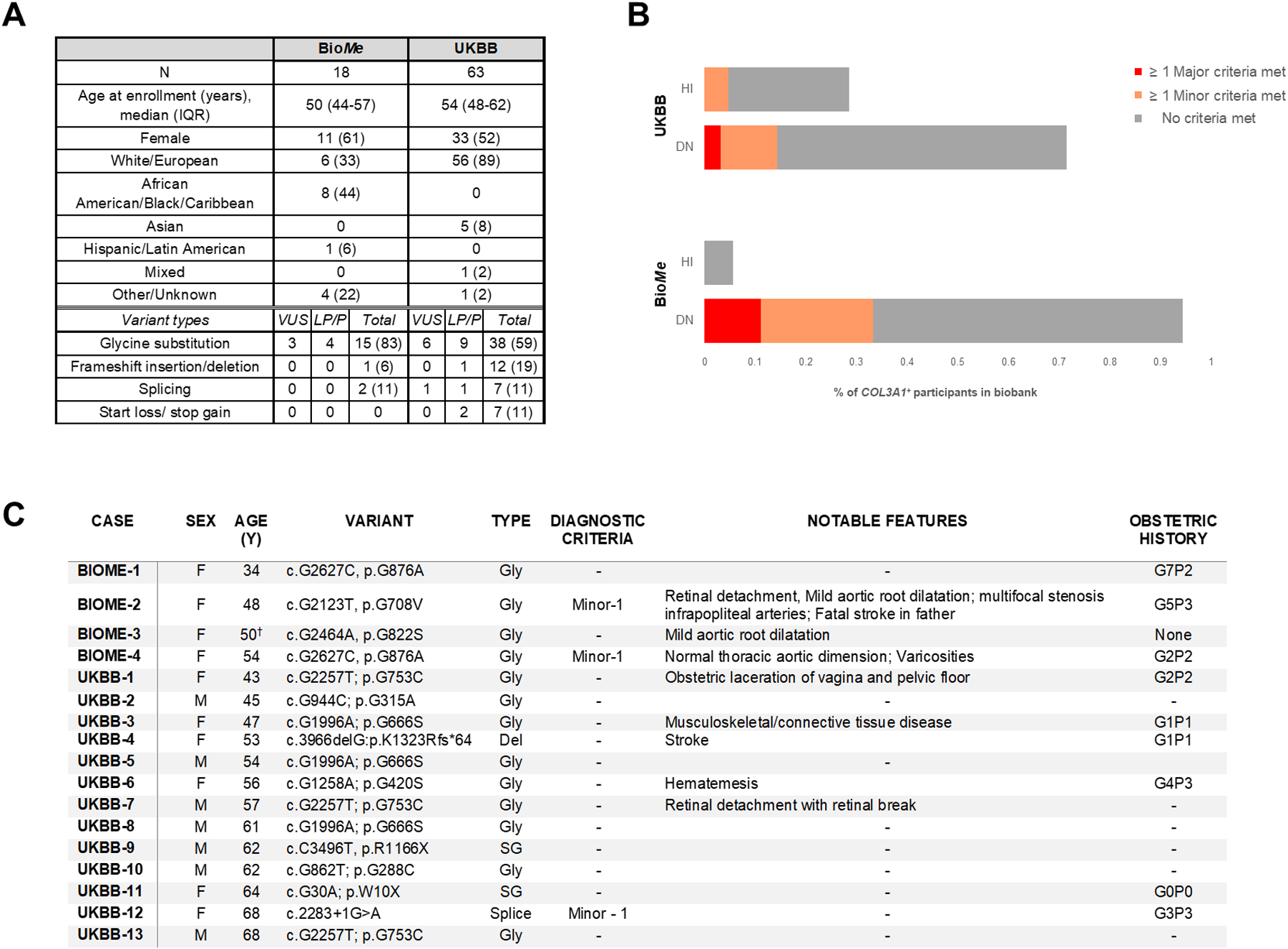
Demographic and clinical data, distribution of variant types and individuals meeting vEDS diagnostic criteria and selection of detailed cases among BioMe and UKBB participants with putatively deleterious COL3A1 variants. A, Demographic data and types of variants for BioMe and UKBB participants with putatively deleterious COL3A1+ genotypes. B, Distribution of vEDS major and minor criteria for BioMe and UKBB participants with putatively deleterious COL3A1 variants stratified according to haplo-insufficiency or dominant-negative variant types.. C, Table of biobank participants harboring COL3A1 variants previously reported in association with vEDS (likely pathogenic or pathogenic designations in ClinVar).A: Table entries are number (%) unless otherwise noted; Designations of variants in ClinVar are listed: VUS- variant of uncertain significance; LP- likely pathogenic; P- pathogenic; total indicates all variants including those not classified in ClinVar; B: HI- “haplo-insufficiency” variant; DN- “dominant negative” variant; C: Age listed is at last EHR entry (BioMe) or first assessment (UKBB); Gly- glycine substitution; SG- stop-gain; Spice- essential splice site mutation; Del- frameshift deletion; † - Participant died at age 50 of septic shock from sternal wound infection following mitral/aortic valve replacement
