Fig. 2 |. Interactions between HIV-1 capsids and NuPODs containing copies of a single nup species.
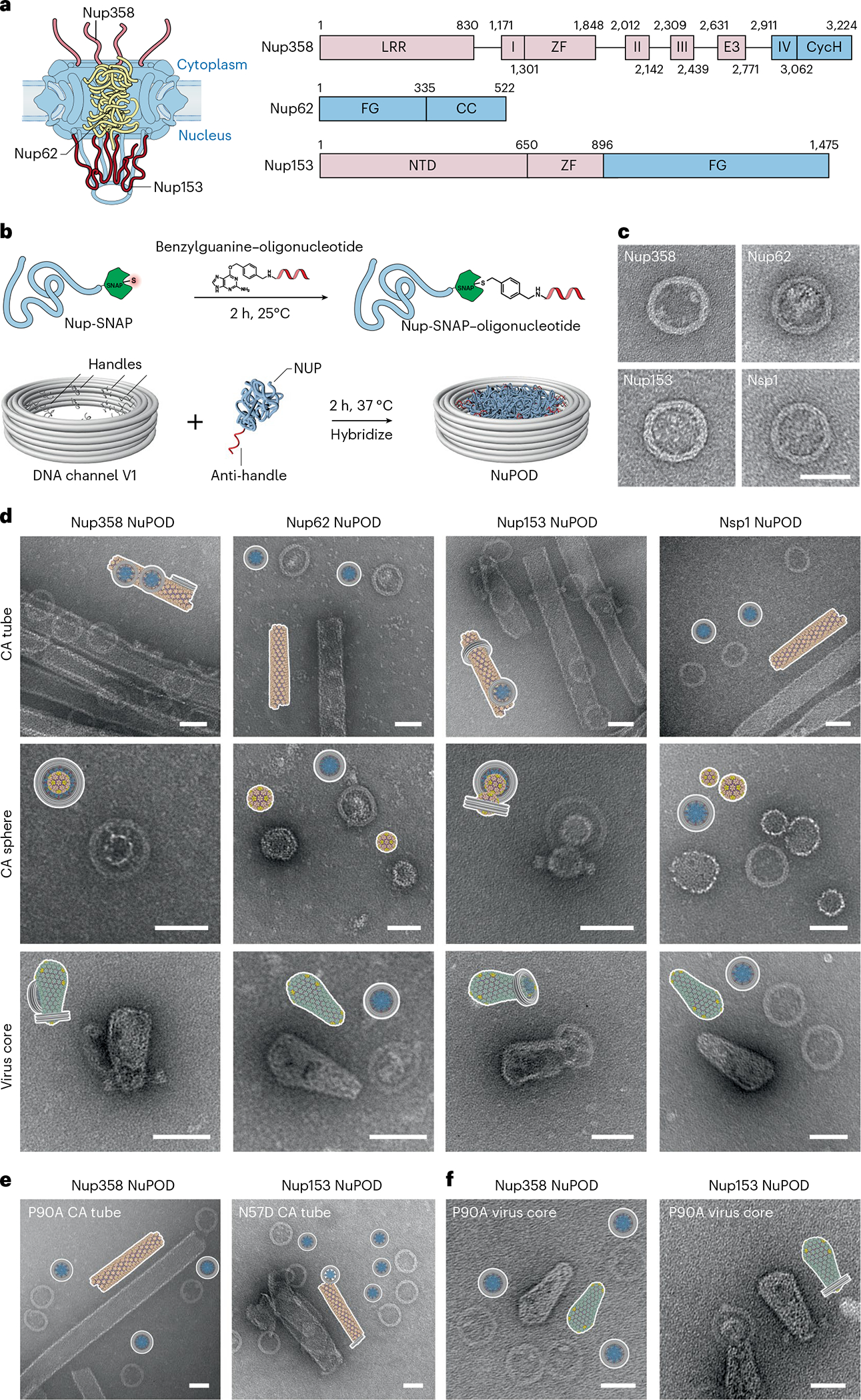
a, Left, a schematic diagram of the organization of capsid-interacting nups in the NPC: Nup358 on the cytoplasmic face, Nup62 in the central channel and Nup153 on the nuclear face. Right, schematic of the protein domains of Nup358, Nup62 and Nup153. LRR (leucine-rich region); Roman numbers I–IV (Ran binding domains I–IV); ZF (zinc finger); E3 (E3 ligase domain); CycH (cyclophilin homology domain); FG (phenylalanine-glycine rich domain); CC (coiled coil domain); NTD (N-terminal domain). The CA-binding regions used in this study are shown in blue. b, Schematic of the conjugation of benzylguanine-labeled anti-handle to SNAP-tag-labeled nup by click chemistry (top) and of attachment of nup–anti-handle conjugates to a DNA-origami channel by DNA hybridization (bottom). c, Negative-stain electron micrographs of various NuPODs. Scale bar, 50 nm. d, Schematics and negative-stain electron micrographs of the Nup358, Nup62, Nup153, and Nsp1 NuPODs (from left to right), mixed with CA tubes (top), CA spheres (middle) and purified virus cores (bottom) that were assembled in vitro. Scale bar, 50 nm. e, Schematics and negative-stain electron micrographs of the Nup358 NuPODs mixed with CA-P90A tubes that were assembled in vitro (left), and Nup153 NuPODs mixed with CA-N57D tubes (right). Scale bar, 50 nm. f, Schematics and negative-stain electron micrographs of the Nup358 (left) and the Nup153 (right) NuPODs mixed with purified virus cores with the P90A mutation. Scale bar, 50 nm. All negative-stain EM experiments were repeated three times with similar results.
