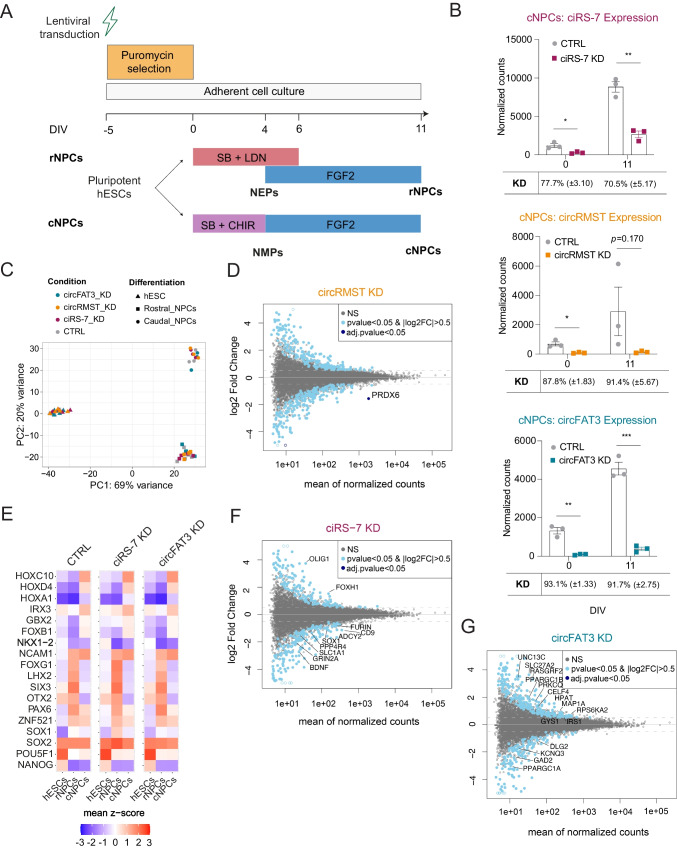
Fig. 2.
Depletion of circFAT3 leads to minor gene expression changes in early neural differentiation. A Pluripotent hESCs were transduced five days prior to differentiation with agoshRNA constructs or empty agoshRNA expression vector as control (CTRL). Transduced cells were selected with puromycin until differentiation onset. For rNPCs, SB and LDN193189 were used from day zero to 6 as well as FGF2 from day 4 to 11. For cNPCs, SB and CHIR were used from day zero to four, and FGF2 from day 4 to 11. Differentiations were performed in biological replicates. B CircRNA quantification using NanoString nCounter technology. ciRS-7, circRMST, and circFAT3 expression as normalized counts in KD versus CTRL cNPCs; n = 3. Mean values per time point ± SEM are shown. Two-tailed unpaired t-tests, ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001. C Principal component analysis (PCA) of CTRL, circRMST KD, circFAT3 KD, and ciRS-7 KD human embryonic stem cells (hESCs; n = 6), rostral neural progenitor cells (rNPCs; n = 4), and caudal NPCs (cNPCs; n = 3). D MA plots depicting global changes in differentially expressed genes (DEGs) in circRMST KD based on DESeq2 analysis. Plotted are average expression across samples and log2 fold changes. E Profiling of days 0 and 11 differentiated H9 hESCs using characteristic markers established in Fig. 1B and as previously described [35]. Normalized DESeq2 counts were log2-transformed (normalized counts + pseudo counts) and converted to a z-scale. F, G MA plots depicting global changes in differentially expressed genes (DEGs) in ciRS-7 KD (F) and circFAT3 KD (G) based on DESeq2 analysis. Plotted are mean expressions against log2 fold changes. KD, knockdown; DIV, days in vitro