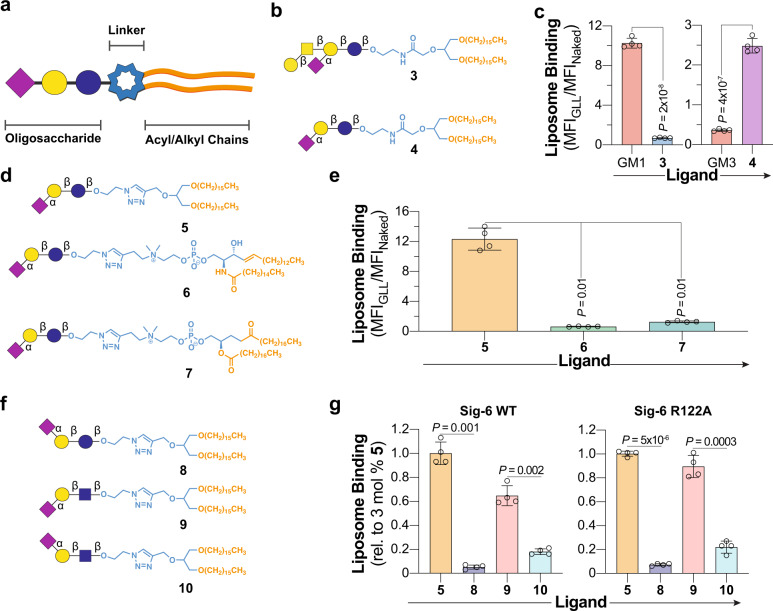
Fig. 3. Exploring the glycolipid binding specificity of Siglec-6 using nGLLs.
a Schematic of a glycolipid structure broken down into three components. b Structures of nGLs 3 and 4 presenting the oligosaccharide of GM1 and GM3, respectively, through an amide-linkage to a 1,3-di-O-hexadecyl glycerol scaffold. c Binding of liposomes formulated with 3 mol% 3 and 4 to WT Siglec-6 CHO cells in the cell assay, relative to liposomes formulated without a ligand (n = 4 technical replicates). d, Structures of nGLs 5, 6, and 7 presenting the oligosaccharide of GM3 triazole-linked to 1,3-di-O-hexadecyl glycerol, phosphatidyl sphingomyelin, and distearoylphosphatidylcholine scaffold, respectively. e Binding of liposomes formulated with nGLs 5, 6, and 7 to WT Siglec-6 CHO cells in the cell assay relative to naked liposomes (n = 4 technical replicates). f Structures of nGLs 8, 9, and 10 presenting an α-(2 → 3)- or α-(2 → 6)-linked sialoside on an underlying lactose or LacNAc core, triazole-linked to 1,3-di-O-hexadecyl glycerol scaffold. g, Binding of liposomes formulated with 5 and 8−10 to WT and R122A Siglec−6 CHO cells in the cell assay relative to liposomes formulated with 3 mol% nGL 5 (n = 4 technical replicates). Data is representative of the mean ± one standard deviation of four technical replicates. For panel c, a two-tailed Student’s t-test was used for statistical analysis. For panels e and g, a Brown–Forsythe and Welch one-way ANOVA was used for statistical analysis.