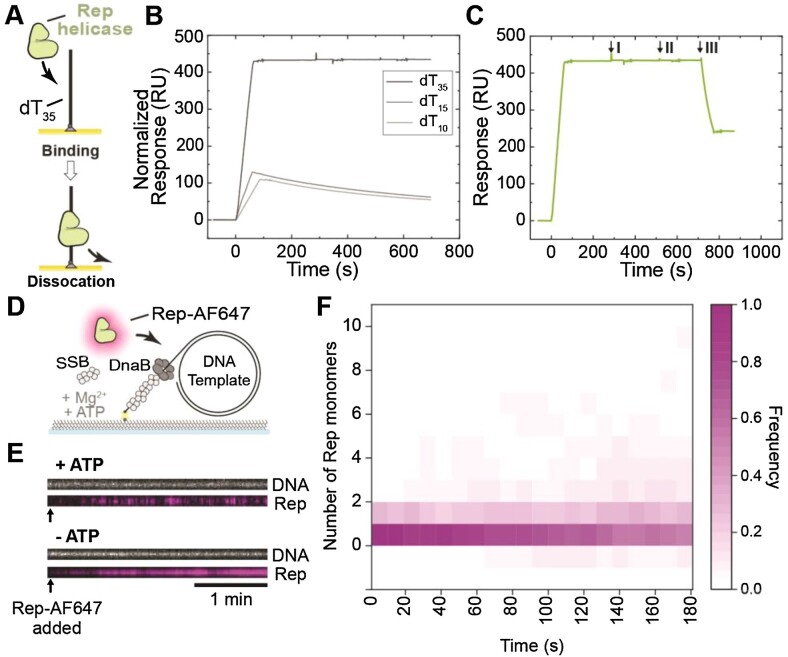
Figure 1.
Visualization of Rep binding to ssDNA. (A) Schematic representation of Rep proteins binding to dT35 oligos in surface plasmon resonance investigations. (B–C) SPR sensorgrams of (B) 20 nM Rep WT association (60 s) and dissociation from dT35 (dark gray), dT15 (gray) and dT10 (light gray). Sensorgrams for Rep binding to dT10 and dT15 were normalized to that for dT35 to enable comparison of binding to the same molar surface density of immobilized oligos. Based on the responses and molecular weights of Rep and oligos, it is estimated that ∼1050 RU of bound Rep corresponds to a stoichiometry n= 1.0 Rep monomers bound to each oligo (Equation 1). (C) Response of dT35-bound Rep WT to interrogation by 200 μM AMP-PNP (I), ADP (II), and ATP (III) injection at 20 μL min–1 for 60 s. Only ATP resulted in fast dissociation of Rep from dT35. (D) Schematic representation of the single-molecule Rep-AF647-DNA binding assay. Rolling-circle DNA templates (2030 bp) are pre-incubated with DnaBC and applied to a microfluidic flow cell. The 5′-biotinylated DNA template couples to the streptavidin functionalized coverslip. Rep-AF647, in the presence of SSB, ATP, and magnesium, is then applied to the flow cell and imaged to monitor for binding. (E) Example kymographs of Rep-AF647 (magenta – bottom) binding to DNA templates (gray – top) in the presence and absence of 5 mM ATP. Arrows indicate the time point of Rep-AF647 addition to the flow cell. (F) Heatmap of the number of Rep-AF647 monomers bound to the DNA template over time in the presence of ATP (n = 65).