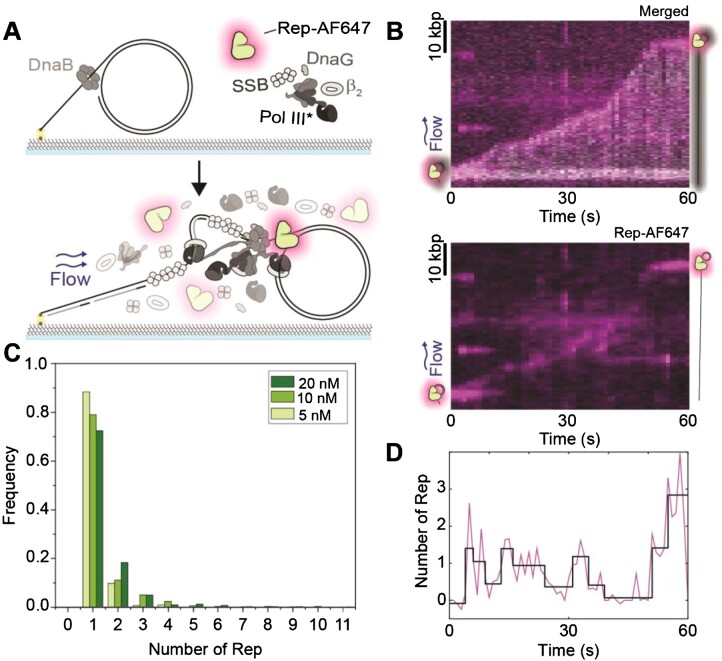
Figure 2.
Rep interacts with processive replisomes. (A) Schematic representation of the single-molecule rolling-circle DNA replication assay in the presence of Rep-AF647. Rolling-circle DNA templates are pre-incubated with DnaBC and immobilized to the flow cell surface. The addition of the replisome components, ATP, NTPs, and dNTPs, results in the initiation of DNA replication. Sytox orange-stained DNA replication products are stretched out by hydrodynamic flow and visualized simultaneously with fluorescent Rep-AF647 proteins. (B) Example kymograph of 20 nM Rep-AF647 interacting with the tip of the Sytox orange stained DNA product (merged – top). The Rep-AF647 intensity alone (bottom) shows its frequent association with and dissociation from the replication fork. (C) Histogram distributions of Rep-AF647 stoichiometry at the replication fork reveal monomeric stoichiometry at 20 nM (dark green; n = 755), 10 nM (green; n= 490) and 5 nM (light green; n= 286). (D) Number of Rep-AF647 as a function of time for the example kymograph in (B) showing variations in Rep stoichiometry at the replication fork during processive replication. Individual steps are detected by change-point analysis (black line).