Figure 3.
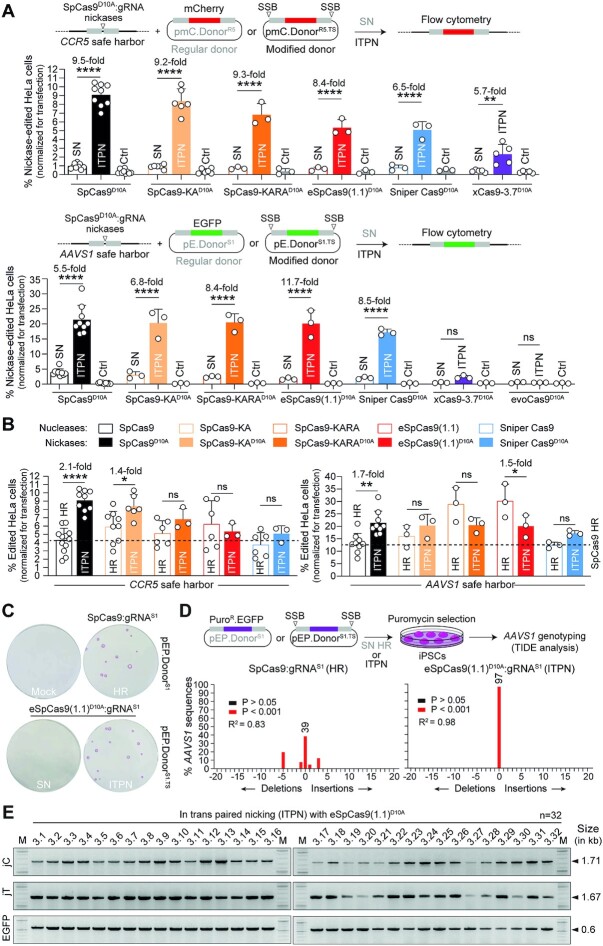
Testing SSB-dependent genome editing using regular versus high-specificity SpCas9D10A nickases. (A) Single nicking and in trans paired nicking genome editing based on high-specificity SpCas9D10A variants. Nickase-dependent genome editing frequencies in HeLa cells transfected with the depicted components targeting CCR5 and AAVS1 were measured by reporter-directed flow cytometry at 17 days post-transfection (top and bottom graphs, respectively). HeLa cells treated with corresponding Cas9D10A nickases and regular donor plasmids in the absence of locus-specific gRNAs served as negative controls. Results are plotted as mean ± SD of at least three independent biological replicates. Significant differences between the indicated datasets were assessed by two-way ANOVA followed by Šidák's multiple comparisons test; ****P < 0.0001, **0.001 < P< 0.01; P> 0.05 was considered non-significant (ns). (B) Comparing standard and in trans paired nicking genome editing strategies at CCR5 and AAVS1. Plotting of datasets presented in panel A corresponding to HeLa cells subjected to nucleases and regular donors or to nickases and target site-modified donors (canonical HR or ITPN strategies, respectively). Dashed lines mark the means of the DSB-dependent genome editing levels obtained with conventional SpCas9 and unmodified HR donor templates. Data are shown as mean ± SD of at least 3 independent biological replicates. Significant differences between the indicated datasets were calculated by two-way ANOVA followed by Šidák's multiple comparisons tests; ****P < 0.0001, **0.001 < P< 0.01, *0.01 < P< 0.05; P> 0.05 was considered non-significant (ns). (C) Testing standard and in trans paired nicking in iPSCs using high-specificity cleaving and nicking CRISPR complexes. iPSCs edited upon exposure to the indicated AAVS1-targeting reagents were selected in the presence of puromycin and the resulting colonies were stained for the pluripotency marker alkaline phosphatase. (D) Probing mutagenic loads in genome-edited iPSCs. iPSCs edited after exposure to the indicated AAVS1-targeting reagents were selected in the presence of puromycin and indel profiles at AAVS1 were examined through tracking of indels by decomposition (TIDE) analysis. (E) Establishing HDR-mediated transgene insertion in iPSCs edited through in trans paired nicking. Junction PCR analysis was performed on randomly picked iPSC clones engineered through pEP.DonorS1.TS and eSpCas9(1.1)D10A:gRNAS1 delivery.