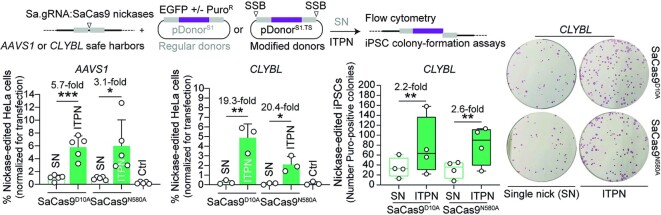
Figure 4.
Genome editing combining regular SN plasmid donors or modified ITPN donors and nicking orthogonal SaCas9 complexes. SaCas9D10A- or SaCas9N580A-dependent genome editing at AAVS1 and CLYBL loci in HeLa cells using EGFP-encoding donors, and at these loci in iPSCs using PuroR.EGFP-encoding donors, was assessed by reporter-directed flow cytometry and colony-formation assays, respectively. The latter assay detected the pluripotency marker alkaline phosphatase to identify puromycin-resistant iPSCs. Controls consisted of cells exposed to regular donor plasmids and nickases lacking locus-specific gRNAs. Data are shown as mean ± SD of at least three independent biological replicates. Significant differences between the indicated datasets were calculated by two-tailed unpaired Student's t tests (left and middle graphs) and two-tailed paired ratio t test (right graph); ***0.0001 < P< 0.001, **0.001 < P< 0.01, *0.01 < P< 0.05.