Figure 8.
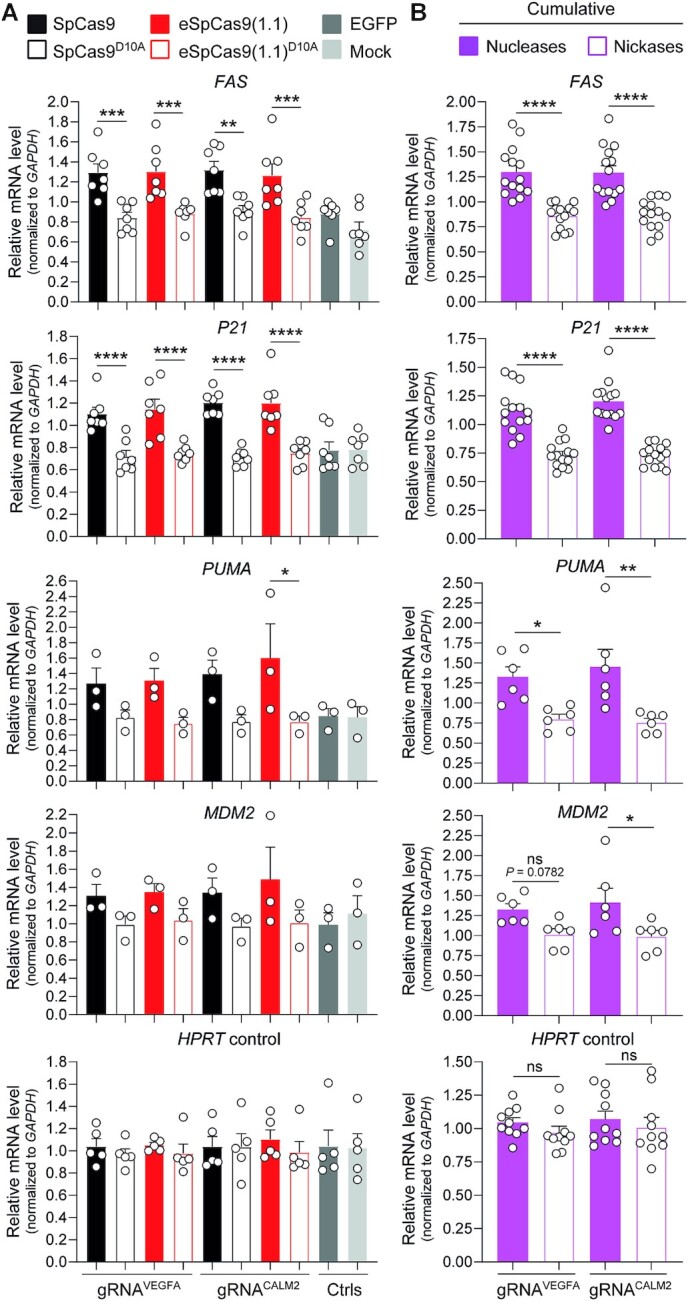
Assessing activation of P53-dependent DNA damage responses in human iPSCs exposed to nucleases versus nickases. (A) Expression analysis of P53 activation-responsive genes. Constructs encoding the indicated Cas9 enzymes and gRNAs conferring high (gRNAVEGFA) or low (gRNACALM2) off-target activities (Supplementary Figure S9), were transfected into iPSCs. RT-qPCR measurements of FAS, P21, PUMA and MDM2 transcripts whose expression is upregulated upon P53 activation (minimum n = 3 independent biological replicates). Targeting HPRT1 transcripts served for RT-qPCR measurements of a housekeeping control gene (n = 5 independent biological replicates). Additional controls consisted of targeting FAS, P21, PUMA, MDM2 and HPRT1 transcripts in mock-transfected iPSCs and in iPSCs transfected with an EGFP-encoding plasmid. Significances were calculated with one-way ANOVA followed by Tukey's test for multiple comparisons; ****P < 0.0001, ***0.0001 < P< 0.001, **0.001 < P< 0.01, *0.01 < P< 0.05. (B) Cumulative comparison of cleaving versus nicking effects on P53-responsive gene modulation. Combined RT-qPCR datasets derived from iPSCs treated with nucleases SpCas9 and eSpCas9(1.1) or nickases SpCas9D10A and eSpCas9(1.1)D10A. Significances were calculated with two-way ANOVA followed by Šidák's test for multiple comparisons; ****P < 0.0001, **0.001 < P< 0.01, *0.01 < P< 0.05; P> 0.05 was considered non-significant (ns).
