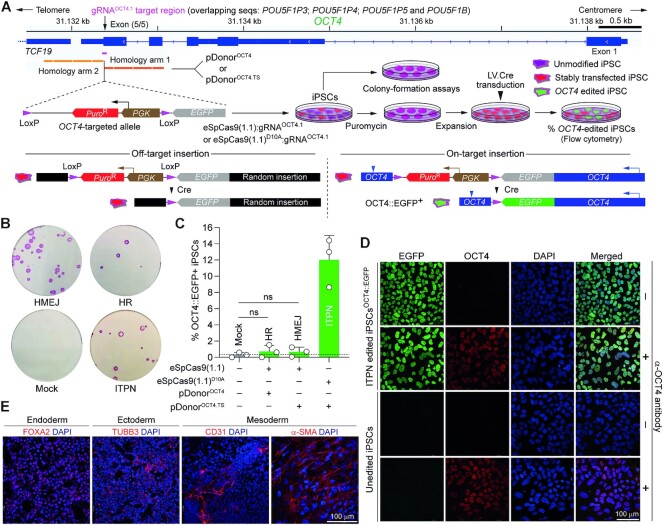
Figure 9.
Testing DSB- versus SSB-dependent genome editing strategies at essential OCT4 alleles in human iPSCs using high-specificity CRISPR complexes. (A) Experimental setup for tracking OCT4 gene editing events. iPSCs exposed to the indicated reagents designed to elicit canonical HR, HMEJ or ITPN were traced by colony-formation assays upon puromycin selection and alkaline phosphatase staining and by a genetic assay reporting live-cell OCT4 gene targeting events upon Cre recombinase delivery. (B) Detection of stably transfected iPSC colonies. Picture of a representative colony-formation assay is shown. (C) Detection of OCT4 gene editing events. The frequencies of OCT4 edited cells (OCT4::EGFP+) in puromycin-resistant iPSC populations were determined by EGFP-directed flow cytometry following transduction with Cre-expressing lentivector particles (20 vector particles per cell). Data are presented as mean ± S.D. of independent biological replicates (n = 3). (D) Confocal microscopy analysis of iPSCs edited at OCT4 through ITPN. OCT4::EGFP-expressing iPSCs engineered through ITPN and Cre delivery (iPSCOCT4::EGFP) were analysed through immunofluorescence microscopy for detecting OCT4 and EGFP, respectively. Nuclei were stained with DAPI. The merge of the three fluorescence signals highlights the nuclear localization of the OCT4::EGFP fusion product. Unedited iPSCs served as negative controls. iPSC and iPSCOCT4::EGFP specimens not incubated with the OCT4-specific primary antibody served as staining controls. (E) Assessing the multi-lineage differentiation capacity of iPSCs edited at OCT4 through ITPN. iPSCsOCT4::EGFP generated by ITPN using high-specificity eSpCas9(1.1)D10A were induced to differentiate into cell lineages corresponding to the three embryonic germ layers, i.e. mesoderm, ectoderm and endoderm. Immunofluorescence microscopy detected the indicated embryonic germ layer-specific markers. Nuclei were stained with DAPI.