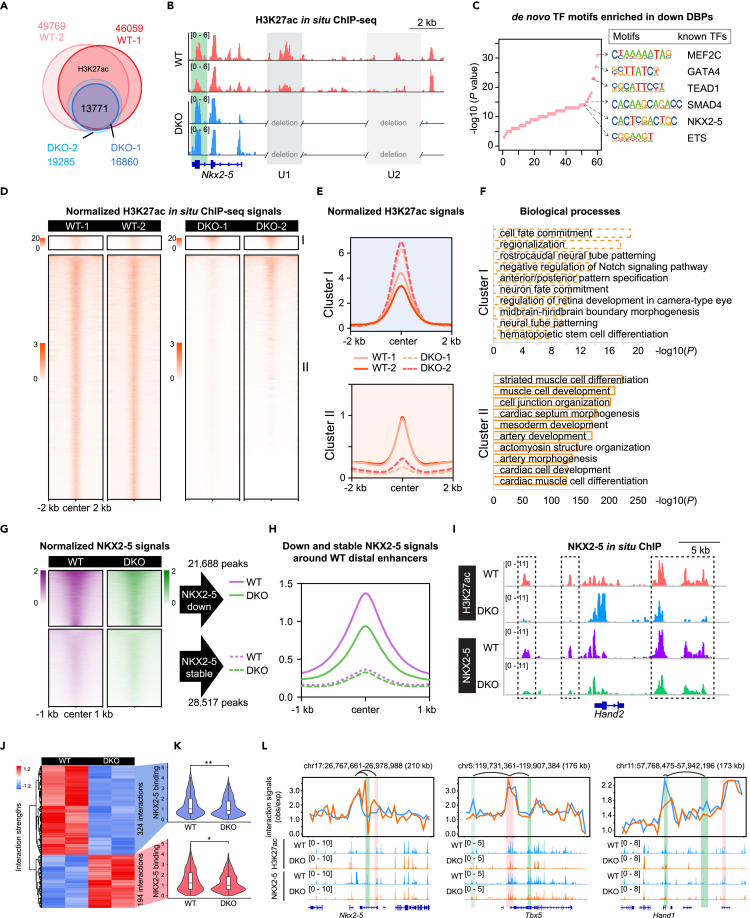
Figure 5.
Genome-wide decommissioning of heart enhancers is associated with perturbation of NKX2-5 binding in DKO mice
(A) Venn diagram showing overlapping of H3K27ac peaks between WT and DKO.
(B) Track view showing H3K27ac in situ ChIP signals in DKO and WT hearts around Nkx2-5 locus. U1 and U2 enhancers were highlighted by light gray, and the highlighted green shading represented increased enhancer signals after U1 and U2 double knockout.
(C) Dot plots showing de novo motifs in downregulated DBPs discovered using Homer. Besides were matched known TFs. p-value was calculated by the Binomial test.
(D) Non-promoter H3K27ac signals around 52,251 non-promoter H3K27ac peaks in E9.5 WT and DKO hearts. Groups consisted of K-means clusters: cluster I (increased; n = 2,046) and cluster II (reduced; n = 50,205). The heatmaps and aggregate plots were both generated with DeepTools.
(E) Aggregate plots of H3K27ac ChIP-seq signals at ± 2 kb of peak centers in (D).
(F) Top 10 GO terms enriched in genes nearby increased and reduced peaks in (D). p-value was calculated by Binomial test.
(G) Heatmap showing NKX2-5 ChIP-seq signals around downregulated H3K27ac peaks in DKO hearts (n = 21,688 for downregulated NKX2-5 peaks and n = 28,517 for stable NKX2-5 peaks).
(H) Aggregation plot of NKX2-5 ChIP-seq signals at ± 1 kb of H3K27ac peak centers in (G).
(I) Track view showing decreased NKX2-5 binding signals at distal enhancers of Hand2 in DKO hearts.
(J) Heatmap displaying the strengths of most interactions between downregulated H3K27ac peaks in E9.5 DKO, and the promoters of heart developmental gene were weakened in DKO. ANOVA was performed for identifying significantly variable interactions.
(K) Violin plot showing the significantly decreased NKX2-5 binding signals on disrupted interactions between enhancers and promoters in (J).
(L) Visualization of chromatin interactions, H3K27ac, and NKX2-5 binding signals around Nkx2-5, Tbx5, and Hand1 loci. The highlighted red and green shading represented increased and decreeased interactions in DKO, respectively. See also Figure S4.