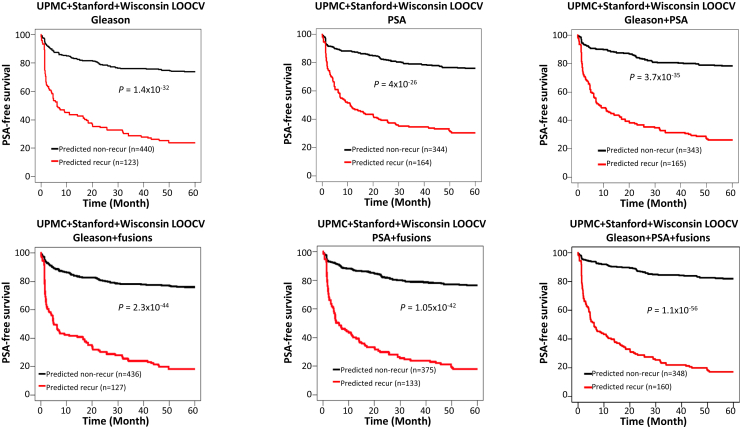
Figure 5.
Fusion gene–containing algorithms enhance PSA-free survival prediction by Gleason score, serum PSA level, or the combination of both, in the combined cohorts of UPMC, Stanford, and Wisconsin. Top panels: Kaplan-Meier analyses of PSA-free survival of prostate cancer patients in the combined cohorts by Gleason (cutoff = 8, left), PSA (cutoff = 9.77 ng/mL, middle), Gleason + PSA (logistic model, right). Bottom panels: Kaplan-Meier analyses of PSA-free survival of prostate cancer patients in the combined cohorts by random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] + Gleason score (left), random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), CLTC-ETV1 (CT ≤ 37), and ACPP-SEC13. (CT ≤ 40)] + PSA (middle), and a random forest model using the detection of five fusion genes [MAN2A1-FER (CT ≤ 34), TRMT11-GRIK2 (CT ≤ 43), MTOR-TP53BP1 (CT ≤ 42), CCNH-C5orf30 (negative), and ACPP-SEC13 (CT ≤ 40)] + Gleason score + PSA (right).