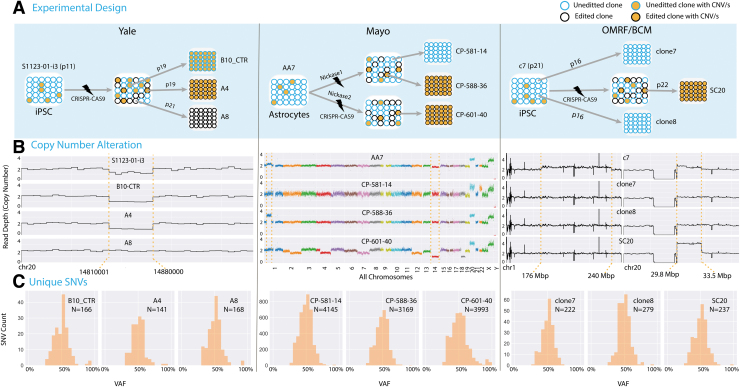
FIG. 1.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR-Cas9) edited clones are not isogenic. (A) In three different experiments conducted by three different groups, CRISPR-Cas9 unedited (in blue circles) and edited (in black circles) single cell clones were analyzed. Orange-filled circles represent clones with CNA. (B) Copy number profiles for the analyzed samples. CNA inherited from unedited parental lines are shown between orange broken lines. In the experiment conducted at Yale, the heterozygous deletion on chromosome 20 was mosaic in the parental unedited iPSC line (90% of cells) [see (A)], and two derived clones (unedited and edited) inherited it. Similarly, in the experiment done at Mayo, multiple mosaic CNA in the parental line (such as duplication on chromosome 1 and deletion of chromosome 14) were inherited by only one out of three clones. In addition, one edited line, CP-601-40, was genomically unstable. In the experiment by OMRF/BCM, a mosaic duplication on chromosome 20 inherited from the unedited parental iPSC line was present in only one of the clones. Clones were selected to avoid mosaic duplication on chromosome 1 present in the parental line. (C) Distribution of private SNVs by allele frequency in each clone (Supplementary Table S1). Clones have hundreds and thousands of point mutations that arose before cloning during culturing of parental lines (see Supplementary Fig. S5 for variant calling). OMRF, Oklahoma Medical Research Foundation; BCM, Baylor College of Medicine; CNA, copy number alteration; iPSC, induced pluripotent stem cell; SNV, single nucleotide variation.