Fig. 3.
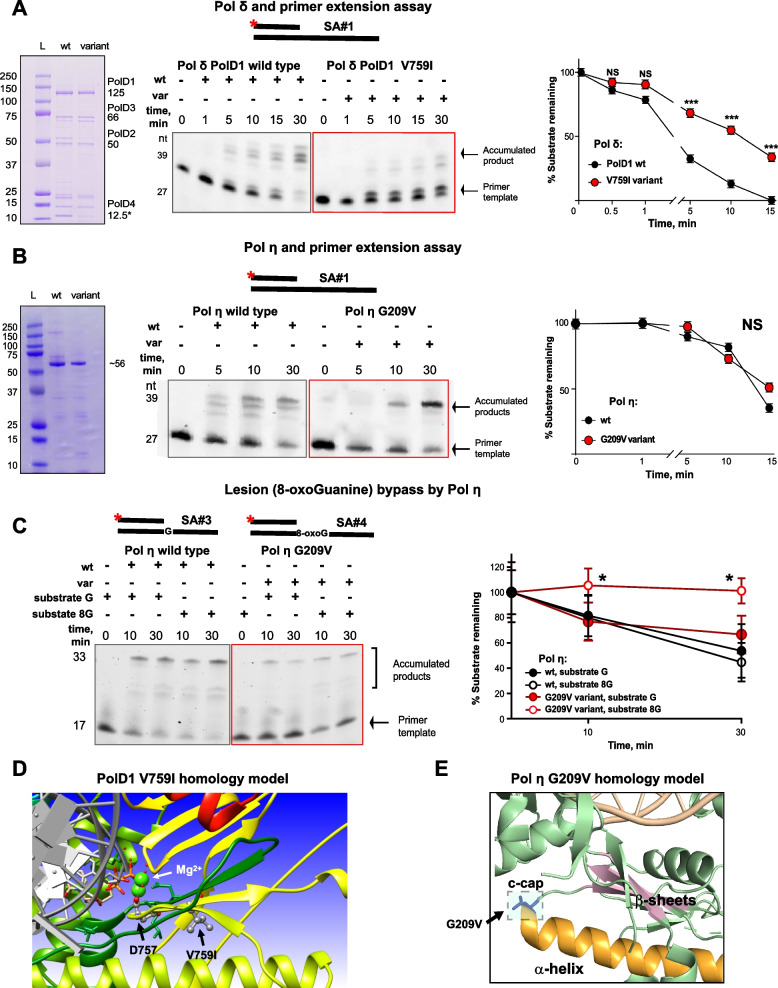
Structural and biochemical assays revealed altered enzymatic activities of the PolD1 and Pol η variants. A. Pol δ complex and primer extension assay. On the left—representative gel image of purified wt and variant Pol δ protein complexes, containing 4 subunits: PolD1 (125 kDa), PolD2 (50 kDa), PolD3 (66 kDa) and PolD4 (12.5 kDa). Center and right – Pol δ complex primer extension assay with quantification. Representative gel image showing reactions performed with 20 nM Cy-3 labeled DNA-duplex template (SA#1), 20 nM of indicated proteins and 500 uM dNTPs. PolD1 V759I complex extended DNA-template less efficiently comparing to wt protein complex. Data for 3 independent repeats are presented. B. Pol η and primer extension assay. On the left—representative gel image of purified wt and variant Pol η catalytic cores (432 amino acids), molecular weight ~ 56 kDa. Center and right – Pol η primer extension assay with quantification. Representative gel image showing reactions performed with 20 nM Cy-3 labeled DNA-duplex template (SA#1), 20 nM of indicated proteins and 500 uM dNTPs. Data for 3 independent repeats is presented. C. Pol η lesion (8-oxoG) bypass assay with quantification. Representative gel image showing reactions performed with 20 nM Cy-3 labeled DNA-duplex template with 8-oxoG in the position, opposite to 3`-OH group (SA#4), and template of the same sequence without lesion (SA#3), 20 nM of indicated proteins and 500 uM dNTPs. Data for 3 independent repeats are presented. For A-C: *** for p < 0.001, ** for p < 0.01, * for p < 0.05 and NS for p > 0.05, unpaired, non-parametric t-test, Mann–Whitney criteria. All template sequences may be found in Supplementary Table 4. D-E. Homology modeling structures using yeast protein templates for human PolD1 V759I (D) and for human Pol η G209V (E)